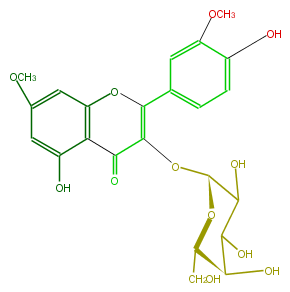
Mol:FL5FCDGA0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.1847 -5.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1847 -5.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4480 -5.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4480 -5.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6676 -4.4416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6676 -4.4416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2548 -3.9495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2548 -3.9495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3779 -4.0610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3779 -4.0610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5976 -4.6646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5976 -4.6646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4744 -3.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4744 -3.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0616 -2.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0616 -2.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5711 -2.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5711 -2.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7908 -3.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7908 -3.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9676 -3.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9676 -3.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9839 -2.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9839 -2.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7599 -1.8582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7599 -1.8582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1808 -1.3568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1808 -1.3568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8255 -1.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8255 -1.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0494 -2.0856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0494 -2.0856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6286 -2.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6286 -2.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4909 -2.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4909 -2.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9416 -0.7694 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.9416 -0.7694 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.5540 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.5540 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.2994 -1.2278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2994 -1.2278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0494 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0494 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4371 -0.7694 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4371 -0.7694 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.6916 -0.9824 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.6916 -0.9824 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.2145 -0.9643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2145 -0.9643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9604 -0.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9604 -0.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2075 0.0876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2075 0.0876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4684 -0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4684 -0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2999 -4.3300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2999 -4.3300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6660 -1.9065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6660 -1.9065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6263 -1.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6263 -1.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7320 -5.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7320 -5.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4980 -6.2589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4980 -6.2589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6199 -1.3985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6199 -1.3985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3344 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3344 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 1 32 1 0 0 0 0 | + | 1 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 22 34 1 0 0 0 0 | + | 22 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 37 | + | M SBL 3 1 37 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 37 2.6199 -1.3985 | + | M SVB 3 37 2.6199 -1.3985 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 35 | + | M SBL 2 1 35 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 35 -3.3344 0.5506 | + | M SVB 2 35 -3.3344 0.5506 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 33 | + | M SBL 1 1 33 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 33 0.6546 1.811 | + | M SVB 1 33 0.6546 1.811 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FCDGA0001 | + | ID FL5FCDGA0001 |
| − | KNApSAcK_ID C00005606 | + | KNApSAcK_ID C00005606 |
| − | NAME Rhamnazin 3-galactoside | + | NAME Rhamnazin 3-galactoside |
| − | CAS_RN 59359-34-1 | + | CAS_RN 59359-34-1 |
| − | FORMULA C23H24O12 | + | FORMULA C23H24O12 |
| − | EXACTMASS 492.126776232 | + | EXACTMASS 492.126776232 |
| − | AVERAGEMASS 492.42946 | + | AVERAGEMASS 492.42946 |
| − | SMILES O(c(c(O)4)cc(cc4)C(O2)=C(C(=O)c(c3O)c2cc(c3)OC)O[C@H](O1)C(O)C([C@H]([C@@H](CO)1)O)O)C | + | SMILES O(c(c(O)4)cc(cc4)C(O2)=C(C(=O)c(c3O)c2cc(c3)OC)O[C@H](O1)C(O)C([C@H]([C@@H](CO)1)O)O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-0.1847 -5.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4480 -5.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6676 -4.4416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2548 -3.9495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3779 -4.0610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5976 -4.6646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4744 -3.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0616 -2.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5711 -2.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7908 -3.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9676 -3.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9839 -2.4734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7599 -1.8582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1808 -1.3568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8255 -1.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0494 -2.0856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6286 -2.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4909 -2.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9416 -0.7694 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.5540 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.2994 -1.2278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0494 -1.4409 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4371 -0.7694 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6916 -0.9824 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.2145 -0.9643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9604 -0.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2075 0.0876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4684 -0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2999 -4.3300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6660 -1.9065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6263 -1.6276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7320 -5.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4980 -6.2589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6199 -1.3985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3344 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
16 30 1 0 0 0 0
30 31 1 0 0 0 0
1 32 1 0 0 0 0
32 33 1 0 0 0 0
22 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 37
M SMT 3 CH2OH
M SVB 3 37 2.6199 -1.3985
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 35
M SMT 2 OCH3
M SVB 2 35 -3.3344 0.5506
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 33
M SMT 1 OCH3
M SVB 1 33 0.6546 1.811
S SKP 8
ID FL5FCDGA0001
KNApSAcK_ID C00005606
NAME Rhamnazin 3-galactoside
CAS_RN 59359-34-1
FORMULA C23H24O12
EXACTMASS 492.126776232
AVERAGEMASS 492.42946
SMILES O(c(c(O)4)cc(cc4)C(O2)=C(C(=O)c(c3O)c2cc(c3)OC)O[C@H](O1)C(O)C([C@H]([C@@H](CO)1)O)O)C
M END