Mol:FL5FCBGL0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0349 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0349 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0349 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0349 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4786 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4786 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9223 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9223 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9223 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9223 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4786 2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4786 2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3660 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3660 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8097 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8097 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8097 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8097 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3660 2.2684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3660 2.2684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3660 0.4829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3660 0.4829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2536 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2536 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3133 1.9410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3133 1.9410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8803 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8803 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8803 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8803 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3133 3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3133 3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2536 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2536 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1616 0.7546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1616 0.7546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8838 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8838 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4962 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4962 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2416 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2416 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9916 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9916 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3793 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3793 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6338 1.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6338 1.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1567 1.0512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1567 1.0512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9026 1.4986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9026 1.4986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8656 1.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8656 1.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4786 0.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4786 0.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6205 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6205 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9661 -0.0240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9661 -0.0240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9257 -0.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9257 -0.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5908 -0.7959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5908 -0.7959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3069 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3069 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3069 -1.9096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3069 -1.9096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6754 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6754 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1119 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1119 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5483 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5483 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5483 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5483 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1119 -3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1119 -3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6754 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6754 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0150 -3.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0150 -3.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5908 2.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5908 2.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7939 2.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7939 2.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4471 3.2503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4471 3.2503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1616 2.8378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1616 2.8378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 3 28 1 0 0 0 0 | + | 3 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 1 42 1 0 0 0 0 | + | 1 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 44 1 0 0 0 0 | + | 15 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 46 -5.9998 7.9947 | + | M SBV 1 46 -5.9998 7.9947 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 -4.8771 7.7401 | + | M SBV 2 48 -4.8771 7.7401 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FCBGL0002 | + | ID FL5FCBGL0002 |
| − | KNApSAcK_ID C00005933 | + | KNApSAcK_ID C00005933 |
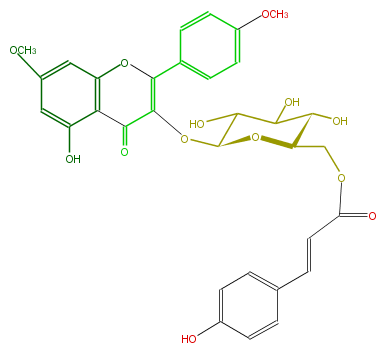
| − | NAME Kaempferol 7,4'-dimethyl ether 3-(6''-(E)-p-coumarylglucoside) | + | NAME Kaempferol 7,4'-dimethyl ether 3-(6''-(E)-p-coumarylglucoside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C32H30O13 | + | FORMULA C32H30O13 |
| − | EXACTMASS 622.168641046 | + | EXACTMASS 622.168641046 |
| − | AVERAGEMASS 622.5728 | + | AVERAGEMASS 622.5728 |
| − | SMILES OC(C(O)2)C(OC(C4=O)=C(c(c5)ccc(c5)OC)Oc(c43)cc(cc3O)OC)OC(C2O)COC(C=Cc(c1)ccc(c1)O)=O | + | SMILES OC(C(O)2)C(OC(C4=O)=C(c(c5)ccc(c5)OC)Oc(c43)cc(cc3O)OC)OC(C2O)COC(C=Cc(c1)ccc(c1)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-3.0349 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0349 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4786 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9223 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9223 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4786 2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3660 0.9837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8097 1.3049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8097 1.9473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3660 2.2684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3660 0.4829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2536 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3133 1.9410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8803 2.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8803 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3133 3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2536 2.9230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1616 0.7546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8838 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4962 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2416 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9916 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3793 1.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6338 1.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1567 1.0512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9026 1.4986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8656 1.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4786 0.3416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6205 0.5746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9661 -0.0240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9257 -0.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5908 -0.7959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3069 -1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3069 -1.9096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6754 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1119 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5483 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5483 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1119 -3.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6754 -2.9250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0150 -3.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5908 2.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7939 2.3156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4471 3.2503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1616 2.8378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
3 28 1 0 0 0 0
22 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
38 41 1 0 0 0 0
1 42 1 0 0 0 0
42 43 1 0 0 0 0
15 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 ^OCH3
M SBV 1 46 -5.9998 7.9947
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 -4.8771 7.7401
S SKP 8
ID FL5FCBGL0002
KNApSAcK_ID C00005933
NAME Kaempferol 7,4'-dimethyl ether 3-(6''-(E)-p-coumarylglucoside)
CAS_RN -
FORMULA C32H30O13
EXACTMASS 622.168641046
AVERAGEMASS 622.5728
SMILES OC(C(O)2)C(OC(C4=O)=C(c(c5)ccc(c5)OC)Oc(c43)cc(cc3O)OC)OC(C2O)COC(C=Cc(c1)ccc(c1)O)=O
M END