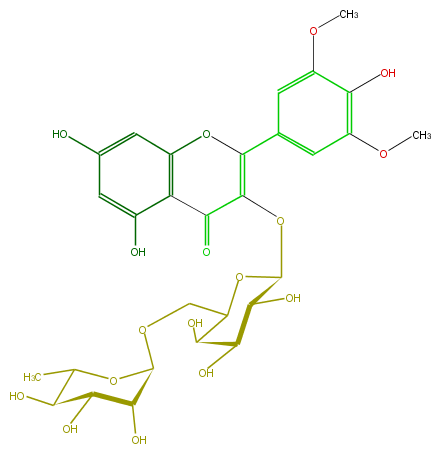
Mol:FL5FAIGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.9005 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9005 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1860 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1860 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1860 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1860 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9005 3.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9005 3.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6150 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6150 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6150 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6150 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4716 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4716 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2428 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2428 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9573 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9573 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9573 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9573 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2428 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2428 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4716 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4716 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6717 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6717 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3862 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3862 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3862 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3862 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6717 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6717 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2428 -0.5046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2428 -0.5046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1007 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1007 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2471 0.1227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2471 0.1227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6717 -0.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6717 -0.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3295 3.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3295 3.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9005 3.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9005 3.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2929 1.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2929 1.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9572 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9572 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5013 4.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5013 4.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4532 -2.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4532 -2.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3711 -2.3625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3711 -2.3625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6264 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6264 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2459 -1.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2459 -1.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4216 -0.9936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4216 -0.9936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1663 -1.7785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1663 -1.7785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5851 -1.5546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5851 -1.5546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5221 -2.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5221 -2.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5376 -1.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5376 -1.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3080 -2.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3080 -2.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4173 -1.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4173 -1.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8969 -2.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8969 -2.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3096 -3.5812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3096 -3.5812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5161 -3.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5161 -3.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7179 -3.5636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7179 -3.5636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3051 -2.8490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3051 -2.8490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0987 -3.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0987 -3.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6605 -4.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6605 -4.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0322 -4.0402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0322 -4.0402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9572 -3.3704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9572 -3.3704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6108 -2.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6108 -2.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 22 25 1 0 0 0 0 | + | 22 25 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 28 36 1 0 0 0 0 | + | 28 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 39 44 1 0 0 0 0 | + | 39 44 1 0 0 0 0 |
| − | 38 45 1 0 0 0 0 | + | 38 45 1 0 0 0 0 |
| − | 37 46 1 0 0 0 0 | + | 37 46 1 0 0 0 0 |
| − | 41 33 1 0 0 0 0 | + | 41 33 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAIGS0005 | + | ID FL5FAIGS0005 |
| − | FORMULA C29H34O17 | + | FORMULA C29H34O17 |
| − | EXACTMASS 654.179599662 | + | EXACTMASS 654.179599662 |
| − | AVERAGEMASS 654.57006 | + | AVERAGEMASS 654.57006 |
| − | SMILES c(C(O2)=C(OC(O4)C(O)C(C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)O)C(=O)c(c(O)3)c2cc(c3)O)(c1)cc(c(c1OC)O)OC | + | SMILES c(C(O2)=C(OC(O4)C(O)C(C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)O)C(=O)c(c(O)3)c2cc(c3)O)(c1)cc(c(c1OC)O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
1.9005 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1860 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1860 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9005 3.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6150 2.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6150 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4716 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2428 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9573 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9573 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2428 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4716 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6717 1.8523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3862 1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3862 0.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6717 0.2024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2428 -0.5046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1007 1.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2471 0.1227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6717 -0.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3295 3.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9005 3.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2929 1.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9572 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5013 4.2615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4532 -2.3236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3711 -2.3625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6264 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2459 -1.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4216 -0.9936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1663 -1.7785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5851 -1.5546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5221 -2.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5376 -1.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3080 -2.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4173 -1.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8969 -2.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3096 -3.5812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5161 -3.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7179 -3.5636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3051 -2.8490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0987 -3.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6605 -4.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0322 -4.0402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9572 -3.3704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6108 -2.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
6 23 1 0 0 0 0
23 24 1 0 0 0 0
22 25 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
26 34 1 0 0 0 0
27 35 1 0 0 0 0
28 36 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
39 44 1 0 0 0 0
38 45 1 0 0 0 0
37 46 1 0 0 0 0
41 33 1 0 0 0 0
29 19 1 0 0 0 0
S SKP 5
ID FL5FAIGS0005
FORMULA C29H34O17
EXACTMASS 654.179599662
AVERAGEMASS 654.57006
SMILES c(C(O2)=C(OC(O4)C(O)C(C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)O)C(=O)c(c(O)3)c2cc(c3)O)(c1)cc(c(c1OC)O)OC
M END