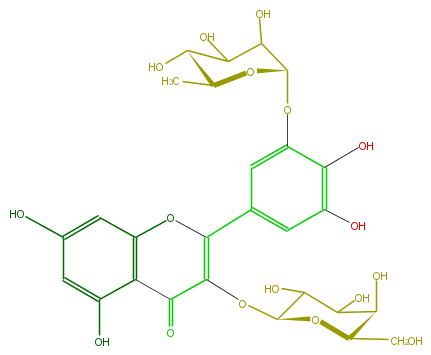
Mol:FL5FAGGA0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2545 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2545 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2545 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2545 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5400 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5400 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8255 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8255 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8255 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8255 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5400 -0.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5400 -0.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1110 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1110 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3965 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3965 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3965 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3965 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1110 -0.4423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1110 -0.4423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1110 -2.7356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1110 -2.7356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5031 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5031 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2314 -0.7040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2314 -0.7040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9597 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9597 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9597 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9597 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2314 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2314 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5031 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5031 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5400 -2.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5400 -2.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7390 1.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7390 1.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1080 -0.3621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1080 -0.3621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3402 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3402 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2280 1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2280 1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5653 -0.6021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5653 -0.6021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5606 -1.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5606 -1.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0142 -2.4833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0142 -2.4833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7869 -2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7869 -2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4538 -2.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4538 -2.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0004 -2.3226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0004 -2.3226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2277 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2277 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9841 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9841 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6499 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6499 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0084 -1.5896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0084 -1.5896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7821 2.8864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7821 2.8864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2561 2.1923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2561 2.1923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5010 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5010 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2317 2.4946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2317 2.4946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 3.0256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 3.0256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0383 2.6760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0383 2.6760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3230 2.5885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3230 2.5885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9921 2.2953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9921 2.2953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4562 3.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4562 3.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6651 3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6651 3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3588 -2.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3588 -2.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1080 -3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1080 -3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 29 31 1 0 0 0 0 | + | 29 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 36 22 1 0 0 0 0 | + | 36 22 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 27 43 1 0 0 0 0 | + | 27 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 48 -0.9050 0.0265 | + | M SBV 1 48 -0.9050 0.0265 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAGGA0004 | + | ID FL5FAGGA0004 |
| − | FORMULA C27H30O17 | + | FORMULA C27H30O17 |
| − | EXACTMASS 626.148299534 | + | EXACTMASS 626.148299534 |
| − | AVERAGEMASS 626.5169000000001 | + | AVERAGEMASS 626.5169000000001 |
| − | SMILES C(C(C5O)OC(C(O)C(O)5)OC(=C2c(c3)cc(OC(O4)C(C(C(C4C)O)O)O)c(O)c3O)C(=O)c(c1O)c(O2)cc(c1)O)O | + | SMILES C(C(C5O)OC(C(O)C(O)5)OC(=C2c(c3)cc(OC(O4)C(C(C(C4C)O)O)O)c(O)c3O)C(=O)c(c1O)c(O2)cc(c1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.2545 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2545 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5400 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8255 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8255 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5400 -0.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1110 -2.0924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3965 -1.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3965 -0.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1110 -0.4423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1110 -2.7356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5031 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2314 -0.7040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9597 -0.2835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9597 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2314 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5031 0.5574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5400 -2.9171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7390 1.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1080 -0.3621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3402 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2280 1.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5653 -0.6021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5606 -1.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0142 -2.4833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7869 -2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4538 -2.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0004 -2.3226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2277 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9841 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6499 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0084 -1.5896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7821 2.8864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2561 2.1923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5010 2.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2317 2.4946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 3.0256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0383 2.6760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3230 2.5885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9921 2.2953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4562 3.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6651 3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3588 -2.8955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1080 -3.6448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
14 23 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 27 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
24 30 1 0 0 0 0
29 31 1 0 0 0 0
28 32 1 0 0 0 0
25 21 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
38 41 1 0 0 0 0
36 22 1 0 0 0 0
37 42 1 0 0 0 0
43 44 1 0 0 0 0
27 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 48
M SMT 1 CH2OH
M SBV 1 48 -0.9050 0.0265
S SKP 5
ID FL5FAGGA0004
FORMULA C27H30O17
EXACTMASS 626.148299534
AVERAGEMASS 626.5169000000001
SMILES C(C(C5O)OC(C(O)C(O)5)OC(=C2c(c3)cc(OC(O4)C(C(C(C4C)O)O)O)c(O)c3O)C(=O)c(c1O)c(O2)cc(c1)O)O
M END