Mol:FL5FADGS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4547 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4547 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4547 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4547 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1016 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1016 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6579 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6579 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6579 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6579 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1016 0.4818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1016 0.4818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2142 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2142 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7705 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7705 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7705 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7705 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2142 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2142 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2142 -1.3038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2142 -1.3038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4710 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4710 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0380 0.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0380 0.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6049 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6049 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6049 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6049 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0380 1.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0380 1.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4710 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4710 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1016 -1.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1016 -1.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6049 1.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6049 1.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9497 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9497 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3338 -0.8912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3338 -0.8912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3531 0.2721 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3531 0.2721 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9262 -0.2914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9262 -0.2914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3114 -0.0523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3114 -0.0523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7182 -0.0460 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7182 -0.0460 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1492 0.3852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1492 0.3852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7772 0.1597 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.7772 0.1597 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.8303 0.5477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8303 0.5477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5871 -0.2077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5871 -0.2077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9591 -0.6437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9591 -0.6437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2118 -1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2118 -1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5580 -1.2478 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.5580 -1.2478 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.1311 -1.8113 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.1311 -1.8113 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5163 -1.5722 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5163 -1.5722 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9231 -1.5658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9231 -1.5658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3542 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3542 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9822 -1.3602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9822 -1.3602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6488 -1.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6488 -1.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1641 -2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1641 -2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0274 -0.9617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0274 -0.9617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8242 -0.7482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8242 -0.7482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0125 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0125 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3429 1.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3429 1.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3214 2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3214 2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7628 3.0609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7628 3.0609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 27 42 1 0 0 0 0 | + | 27 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 46 -3.0125 0.8855 | + | M SVB 3 46 -3.0125 0.8855 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 44 -3.0274 -0.9617 | + | M SVB 2 44 -3.0274 -0.9617 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 48 3.3214 2.1636 | + | M SVB 1 48 3.3214 2.1636 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGS0016 | + | ID FL5FADGS0016 |
| − | KNApSAcK_ID C00005563 | + | KNApSAcK_ID C00005563 |
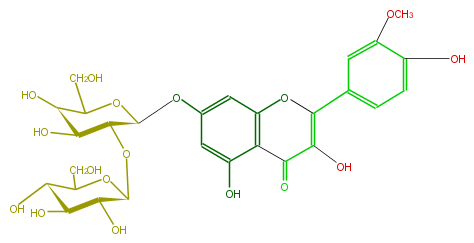
| − | NAME Isorhamnetin 7-sophoroside | + | NAME Isorhamnetin 7-sophoroside |
| − | CAS_RN 123131-39-5 | + | CAS_RN 123131-39-5 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES O(c(c5)cc(c3c(O)5)OC(c(c4)cc(c(O)c4)OC)=C(C3=O)O)[C@@H]([C@H]1O[C@@H]([C@H]2O)OC([C@@H]([C@@H]2O)O)CO)OC([C@@H]([C@H](O)1)O)CO | + | SMILES O(c(c5)cc(c3c(O)5)OC(c(c4)cc(c(O)c4)OC)=C(C3=O)O)[C@@H]([C@H]1O[C@@H]([C@H]2O)OC([C@@H]([C@@H]2O)O)CO)OC([C@@H]([C@H](O)1)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-0.4547 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4547 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1016 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6579 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6579 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1016 0.4818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2142 -0.8029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7705 -0.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7705 0.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2142 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2142 -1.3038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4710 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0380 0.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6049 0.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6049 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0380 1.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4710 1.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1016 -1.4450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6049 1.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9497 0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3338 -0.8912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3531 0.2721 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9262 -0.2914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3114 -0.0523 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7182 -0.0460 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1492 0.3852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7772 0.1597 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.8303 0.5477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5871 -0.2077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9591 -0.6437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2118 -1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5580 -1.2478 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.1311 -1.8113 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5163 -1.5722 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9231 -1.5658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3542 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9822 -1.3602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6488 -1.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1641 -2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0274 -0.9617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8242 -0.7482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0125 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3429 1.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3214 2.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7628 3.0609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
31 32 1 0 0 0 0
35 30 1 0 0 0 0
25 20 1 0 0 0 0
37 40 1 0 0 0 0
40 41 1 0 0 0 0
27 42 1 0 0 0 0
42 43 1 0 0 0 0
16 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 46
M SMT 3 CH2OH
M SVB 3 46 -3.0125 0.8855
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 CH2OH
M SVB 2 44 -3.0274 -0.9617
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 48
M SMT 1 OCH3
M SVB 1 48 3.3214 2.1636
S SKP 8
ID FL5FADGS0016
KNApSAcK_ID C00005563
NAME Isorhamnetin 7-sophoroside
CAS_RN 123131-39-5
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES O(c(c5)cc(c3c(O)5)OC(c(c4)cc(c(O)c4)OC)=C(C3=O)O)[C@@H]([C@H]1O[C@@H]([C@H]2O)OC([C@@H]([C@@H]2O)O)CO)OC([C@@H]([C@H](O)1)O)CO
M END