Mol:FL5FADGS0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 34 37 0 0 0 0 0 0 0 0999 V2000 | + | 34 37 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0282 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0282 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0282 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0282 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4719 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4719 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0844 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0844 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0844 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0844 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4719 0.7848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4719 0.7848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6407 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6407 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1970 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1970 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1970 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1970 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6407 0.7848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6407 0.7848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6407 -1.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6407 -1.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7531 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7531 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3201 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3201 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8871 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8871 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8871 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8871 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3201 1.7667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3201 1.7667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7531 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7531 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5843 0.7847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5843 0.7847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4719 -1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4719 -1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8871 1.4394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8871 1.4394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7531 -0.4998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7531 -0.4998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7589 -1.3938 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.7589 -1.3938 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.3165 -1.9778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3165 -1.9778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6795 -1.7301 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6795 -1.7301 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0647 -1.7234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0647 -1.7234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5114 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5114 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0688 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.0688 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.3714 -1.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3714 -1.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0062 -2.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0062 -2.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3144 -2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3144 -2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6035 2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6035 2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0449 3.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0449 3.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0975 -1.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0975 -1.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8452 -0.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8452 -0.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 16 31 1 0 0 0 0 | + | 16 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 33 34 | + | M SAL 2 2 33 34 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 36 -2.1682 -1.1724 | + | M SVB 2 36 -2.1682 -1.1724 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 31 32 | + | M SAL 1 2 31 32 |
| − | M SBL 1 1 34 | + | M SBL 1 1 34 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 34 2.6035 2.3428 | + | M SVB 1 34 2.6035 2.3428 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGS0007 | + | ID FL5FADGS0007 |
| − | KNApSAcK_ID C00005529 | + | KNApSAcK_ID C00005529 |
| − | NAME Isorhamnetin 5-glucoside | + | NAME Isorhamnetin 5-glucoside |
| − | CAS_RN 34199-20-7 | + | CAS_RN 34199-20-7 |
| − | FORMULA C22H22O12 | + | FORMULA C22H22O12 |
| − | EXACTMASS 478.111126168 | + | EXACTMASS 478.111126168 |
| − | AVERAGEMASS 478.40288000000004 | + | AVERAGEMASS 478.40288000000004 |
| − | SMILES O(c(c1)c(O)ccc1C(=C4O)Oc(c2)c(C4=O)c(O[C@@H]([C@@H](O)3)OC(CO)[C@@H]([C@@H]3O)O)cc2O)C | + | SMILES O(c(c1)c(O)ccc1C(=C4O)Oc(c2)c(C4=O)c(O[C@@H]([C@@H](O)3)OC(CO)[C@@H]([C@@H]3O)O)cc2O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
34 37 0 0 0 0 0 0 0 0999 V2000
-1.0282 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0282 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4719 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0844 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0844 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4719 0.7848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6407 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1970 -0.1787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1970 0.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6407 0.7848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6407 -1.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7531 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3201 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8871 0.7847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8871 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3201 1.7667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7531 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5843 0.7847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4719 -1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8871 1.4394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7531 -0.4998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7589 -1.3938 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.3165 -1.9778 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6795 -1.7301 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0647 -1.7234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5114 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0688 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.3714 -1.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0062 -2.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3144 -2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6035 2.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0449 3.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0975 -1.1612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8452 -0.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 19 1 0 0 0 0
16 31 1 0 0 0 0
31 32 1 0 0 0 0
27 33 1 0 0 0 0
33 34 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 33 34
M SBL 2 1 36
M SMT 2 CH2OH
M SVB 2 36 -2.1682 -1.1724
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 31 32
M SBL 1 1 34
M SMT 1 OCH3
M SVB 1 34 2.6035 2.3428
S SKP 8
ID FL5FADGS0007
KNApSAcK_ID C00005529
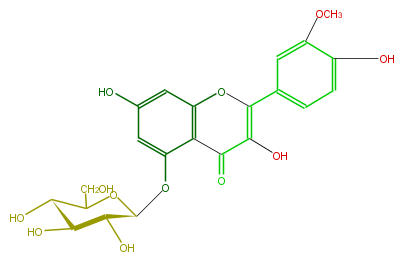
NAME Isorhamnetin 5-glucoside
CAS_RN 34199-20-7
FORMULA C22H22O12
EXACTMASS 478.111126168
AVERAGEMASS 478.40288000000004
SMILES O(c(c1)c(O)ccc1C(=C4O)Oc(c2)c(C4=O)c(O[C@@H]([C@@H](O)3)OC(CO)[C@@H]([C@@H]3O)O)cc2O)C
M END