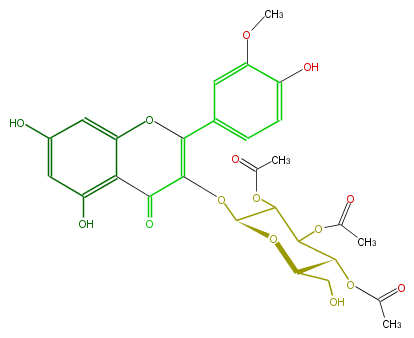
Mol:FL5FADGL0041
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 46 0 0 0 0 0 0 0 0999 V2000 | + | 43 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5619 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5619 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8953 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8953 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8953 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8953 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5619 0.9784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5619 0.9784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2284 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2284 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2284 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2284 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2287 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2287 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5622 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5622 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5622 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5622 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2287 0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2287 0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0685 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0685 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7159 0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7159 0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3633 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3633 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3633 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3633 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7159 2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7159 2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0685 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0685 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9257 2.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9257 2.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7159 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7159 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1384 3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1384 3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8070 0.9276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8070 0.9276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5619 -1.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5619 -1.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2287 -1.0902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2287 -1.0902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2043 -0.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2043 -0.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2782 -0.7898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2782 -0.7898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4980 -1.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4980 -1.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2452 -1.4077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2452 -1.4077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7097 -2.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7097 -2.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4901 -1.8215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4901 -1.8215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7429 -1.4715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7429 -1.4715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3432 -2.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3432 -2.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4614 -2.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4614 -2.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7905 -2.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7905 -2.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3650 -2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3650 -2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8070 -2.3799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8070 -2.3799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4946 -3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4946 -3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1618 -1.2096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1618 -1.2096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5785 -1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5785 -1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7917 -0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7917 -0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0118 -1.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0118 -1.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9057 -0.5822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9057 -0.5822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9057 -0.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9057 -0.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4917 0.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4917 0.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2885 0.1881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2885 0.1881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 7 22 2 0 0 0 0 | + | 7 22 2 0 0 0 0 |
| − | 8 23 1 0 0 0 0 | + | 8 23 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 29 36 1 0 0 0 0 | + | 29 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 24 40 1 0 0 0 0 | + | 24 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0041 | + | ID FL5FADGL0041 |
| − | KNApSAcK_ID C00011133 | + | KNApSAcK_ID C00011133 |
| − | NAME Isorhamnetin 3-O-beta-D-2'',3'',4''-triacetylglucopyranoside;5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[(2,3,4-tri-O-acetyl-beta-D-glucopyranosyl)oxy]-4H-1-benzopyran-4-one | + | NAME Isorhamnetin 3-O-beta-D-2'',3'',4''-triacetylglucopyranoside;5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[(2,3,4-tri-O-acetyl-beta-D-glucopyranosyl)oxy]-4H-1-benzopyran-4-one |
| − | CAS_RN 600736-88-7 | + | CAS_RN 600736-88-7 |
| − | FORMULA C28H28O15 | + | FORMULA C28H28O15 |
| − | EXACTMASS 604.1428202259999 | + | EXACTMASS 604.1428202259999 |
| − | AVERAGEMASS 604.51292 | + | AVERAGEMASS 604.51292 |
| − | SMILES c(c1C(O3)=C(C(=O)c(c4O)c(cc(O)c4)3)OC(O2)C(OC(C)=O)C(C(OC(C)=O)C2CO)OC(C)=O)c(OC)c(O)cc1 | + | SMILES c(c1C(O3)=C(C(=O)c(c4O)c(cc(O)c4)3)OC(O2)C(OC(C)=O)C(C(OC(C)=O)C2CO)OC(C)=O)c(OC)c(O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 46 0 0 0 0 0 0 0 0999 V2000
-2.5619 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8953 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8953 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5619 0.9784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2284 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2284 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2287 -0.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5622 -0.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5622 0.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2287 0.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0685 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7159 0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3633 0.9577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3633 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7159 2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0685 1.7052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9257 2.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7159 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1384 3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8070 0.9276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5619 -1.1065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2287 -1.0902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2043 -0.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2782 -0.7898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4980 -1.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2452 -1.4077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7097 -2.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4901 -1.8215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7429 -1.4715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3432 -2.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4614 -2.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7905 -2.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3650 -2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8070 -2.3799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4946 -3.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1618 -1.2096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5785 -1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7917 -0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0118 -1.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9057 -0.5822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9057 -0.0330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4917 0.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2885 0.1881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
15 18 1 0 0 0 0
18 19 1 0 0 0 0
5 20 1 0 0 0 0
1 21 1 0 0 0 0
7 22 2 0 0 0 0
8 23 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
23 25 1 0 0 0 0
27 30 1 0 0 0 0
30 31 1 0 0 0 0
28 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
29 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
37 39 1 0 0 0 0
24 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
S SKP 8
ID FL5FADGL0041
KNApSAcK_ID C00011133
NAME Isorhamnetin 3-O-beta-D-2'',3'',4''-triacetylglucopyranoside;5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[(2,3,4-tri-O-acetyl-beta-D-glucopyranosyl)oxy]-4H-1-benzopyran-4-one
CAS_RN 600736-88-7
FORMULA C28H28O15
EXACTMASS 604.1428202259999
AVERAGEMASS 604.51292
SMILES c(c1C(O3)=C(C(=O)c(c4O)c(cc(O)c4)3)OC(O2)C(OC(C)=O)C(C(OC(C)=O)C2CO)OC(C)=O)c(OC)c(O)cc1
M END