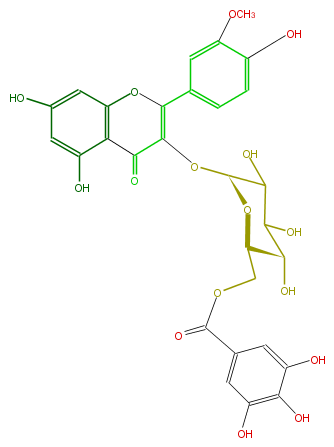
Mol:FL5FADGL0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6883 -3.8001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6883 -3.8001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0678 -3.9664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0678 -3.9664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6136 -3.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6136 -3.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7799 -2.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7799 -2.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4004 -2.7254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4004 -2.7254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8546 -3.1796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8546 -3.1796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3256 -2.4375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3256 -2.4375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4919 -1.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4919 -1.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1124 -1.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1124 -1.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5667 -2.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5667 -2.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8420 -2.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8420 -2.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2786 -1.0305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2786 -1.0305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8157 -0.5675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8157 -0.5675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9851 0.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9851 0.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6174 0.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6174 0.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0804 -0.2286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0804 -0.2286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9109 -0.8610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9109 -0.8610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7927 -1.3334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7927 -1.3334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0609 -0.0732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.0609 -0.0732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.4485 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4485 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2970 -0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2970 -0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0469 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.0469 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.4347 -0.0732 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4347 -0.0732 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.6892 -0.2861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.6892 -0.2861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.7879 -0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7879 -0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9580 0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9580 0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0489 -0.2378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0489 -0.2378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8762 1.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8762 1.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9934 -3.6782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9934 -3.6782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0382 -4.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0382 -4.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6759 -0.7446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6759 -0.7446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7541 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7541 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3729 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3729 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3729 -2.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3729 -2.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0329 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0329 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5521 -1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5521 -1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0713 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0713 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0713 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0713 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5521 -0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5521 -0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0329 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0329 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5521 -0.0181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5521 -0.0181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5905 -0.6176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5905 -0.6176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5905 -1.8167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5905 -1.8167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5636 0.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5636 0.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3160 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3160 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 48 -0.3479 2.5073 | + | M SVB 1 48 -0.3479 2.5073 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0037 | + | ID FL5FADGL0037 |
| − | KNApSAcK_ID C00006008 | + | KNApSAcK_ID C00006008 |
| − | NAME Isorhamnetin 3-(6''-galloylglucoside) | + | NAME Isorhamnetin 3-(6''-galloylglucoside) |
| − | CAS_RN 115651-94-0 | + | CAS_RN 115651-94-0 |
| − | FORMULA C29H26O16 | + | FORMULA C29H26O16 |
| − | EXACTMASS 630.122084784 | + | EXACTMASS 630.122084784 |
| − | AVERAGEMASS 630.5071399999999 | + | AVERAGEMASS 630.5071399999999 |
| − | SMILES [C@@H](OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)(C2O)O[C@@H]([C@H](O)C2O)COC(=O)c(c1)cc(O)c(c(O)1)O | + | SMILES [C@@H](OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)(C2O)O[C@@H]([C@H](O)C2O)COC(=O)c(c1)cc(O)c(c(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-2.6883 -3.8001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0678 -3.9664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6136 -3.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7799 -2.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4004 -2.7254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8546 -3.1796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3256 -2.4375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4919 -1.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1124 -1.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5667 -2.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8420 -2.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2786 -1.0305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8157 -0.5675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9851 0.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6174 0.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0804 -0.2286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9109 -0.8610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7927 -1.3334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0609 -0.0732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.4485 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2970 -0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0469 -0.7446 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4347 -0.0732 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6892 -0.2861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.7879 -0.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9580 0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0489 -0.2378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8762 1.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9934 -3.6782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0382 -4.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6759 -0.7446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7541 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3729 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3729 -2.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0329 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5521 -1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0713 -1.5169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0713 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5521 -0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0329 -0.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5521 -0.0181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5905 -0.6176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5905 -1.8167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5636 0.1942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3160 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
37 43 1 0 0 0 0
16 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 48
M SMT 1 OCH3
M SVB 1 48 -0.3479 2.5073
S SKP 8
ID FL5FADGL0037
KNApSAcK_ID C00006008
NAME Isorhamnetin 3-(6''-galloylglucoside)
CAS_RN 115651-94-0
FORMULA C29H26O16
EXACTMASS 630.122084784
AVERAGEMASS 630.5071399999999
SMILES [C@@H](OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)(C2O)O[C@@H]([C@H](O)C2O)COC(=O)c(c1)cc(O)c(c(O)1)O
M END