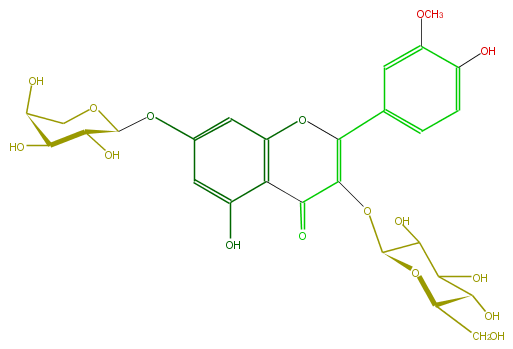
Mol:FL5FADGL0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2161 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2161 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2160 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2160 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4922 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4922 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2315 -0.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2315 -0.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2315 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2315 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4922 0.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4922 0.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9552 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9552 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6789 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6789 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6790 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6790 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9552 0.9463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9552 0.9463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9554 -1.3766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9554 -1.3766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5904 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5904 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3280 0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3280 0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0656 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0656 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0656 1.9588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0656 1.9588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3280 2.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3280 2.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5903 1.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5903 1.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4921 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4921 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5985 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5985 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0804 1.0276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0804 1.0276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2566 -0.8848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2566 -0.8848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2994 -1.5399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2994 -1.5399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5570 -1.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5570 -1.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2289 -2.1121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2289 -2.1121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6053 -2.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6053 -2.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3478 -2.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3478 -2.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6758 -2.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6758 -2.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8822 -1.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8822 -1.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4358 -2.2219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4358 -2.2219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6781 -2.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6781 -2.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5766 1.0493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5766 1.0493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0937 0.4120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0937 0.4120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3982 0.6824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3982 0.6824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7274 0.6896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7274 0.6896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2150 1.1774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2150 1.1774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8233 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8233 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4370 1.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4370 1.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6781 0.3949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6781 0.3949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9149 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9149 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3245 3.0505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3245 3.0505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8987 4.2180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8987 4.2180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4293 -3.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4293 -3.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3409 -4.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3409 -4.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 45 0.0035 -0.6657 | + | M SBV 1 45 0.0035 -0.6657 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 47 -0.8240 0.6002 | + | M SBV 2 47 -0.8240 0.6002 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0012 | + | ID FL5FADGL0012 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(c(C3=O)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2OC)O)O | + | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(c(C3=O)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2OC)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.2161 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2160 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4922 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2315 -0.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2315 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4922 0.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9552 -0.7251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6789 -0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6790 0.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9552 0.9463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9554 -1.3766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5904 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3280 0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0656 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0656 1.9588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3280 2.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5903 1.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4921 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5985 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0804 1.0276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2566 -0.8848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2994 -1.5399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5570 -1.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2289 -2.1121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6053 -2.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3478 -2.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6758 -2.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8822 -1.0806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4358 -2.2219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6781 -2.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5766 1.0493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0937 0.4120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3982 0.6824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7274 0.6896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2150 1.1774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8233 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4370 1.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6781 0.3949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9149 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3245 3.0505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8987 4.2180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4293 -3.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3409 -4.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
21 23 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 20 1 0 0 0 0
40 41 1 0 0 0 0
16 40 1 0 0 0 0
42 43 1 0 0 0 0
25 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 OCH3
M SBV 1 45 0.0035 -0.6657
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 CH2OH
M SBV 2 47 -0.8240 0.6002
S SKP 5
ID FL5FADGL0012
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(c(C3=O)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2OC)O)O
M END