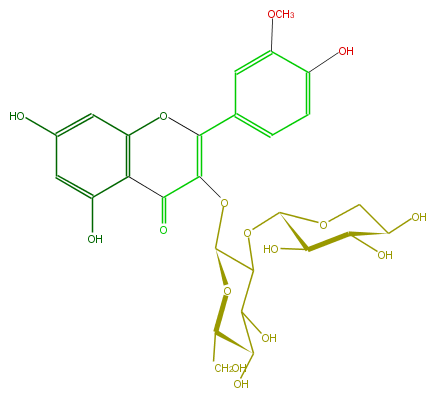
Mol:FL5FADGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2401 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2401 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2401 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2401 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5257 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5257 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8113 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8113 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8113 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8113 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5257 1.0890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5257 1.0890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0968 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0968 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3824 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3824 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3824 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3824 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0968 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0968 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0968 -1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0968 -1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3318 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3318 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0599 0.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0599 0.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7881 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7881 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7881 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7881 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0599 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0599 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3318 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3318 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9543 1.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9543 1.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1238 -0.6478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1238 -0.6478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5257 -1.3855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5257 -1.3855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4891 2.3848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4891 2.3848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6873 -2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6873 -2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0611 -1.5893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0611 -1.5893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1763 -2.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1763 -2.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0611 -3.2563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0611 -3.2563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6873 -3.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6873 -3.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4499 -2.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4499 -2.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5767 -1.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5767 -1.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9408 -3.3660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9408 -3.3660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3788 -4.2822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3788 -4.2822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2113 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2113 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7794 -1.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7794 -1.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5975 -1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5975 -1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3870 -1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3870 -1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8134 -0.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8134 -0.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0975 -1.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0975 -1.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1247 -1.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1247 -1.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2753 -1.6944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2753 -1.6944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9543 -0.9324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9543 -0.9324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1024 -3.9607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1024 -3.9607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3788 -3.8189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3788 -3.8189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0746 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0746 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6415 4.2822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6415 4.2822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.0413 0.7044 | + | M SBV 1 45 0.0413 0.7044 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -0.0147 -0.7797 | + | M SBV 2 47 -0.0147 -0.7797 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0003 | + | ID FL5FADGL0003 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c5O)c(c(C2=O)c(c5)OC(=C2OC(C(OC(C4O)OCC(C4O)O)3)OC(CO)C(C3O)O)c(c1)ccc(c1OC)O)O | + | SMILES c(c5O)c(c(C2=O)c(c5)OC(=C2OC(C(OC(C4O)OCC(C4O)O)3)OC(CO)C(C3O)O)c(c1)ccc(c1OC)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.2401 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2401 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5257 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8113 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8113 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5257 1.0890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0968 -0.5609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3824 -0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3824 0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0968 1.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0968 -1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3318 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0599 0.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7881 1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7881 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0599 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3318 1.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9543 1.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1238 -0.6478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5257 -1.3855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4891 2.3848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6873 -2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0611 -1.5893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1763 -2.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0611 -3.2563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6873 -3.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4499 -2.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5767 -1.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9408 -3.3660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3788 -4.2822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2113 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7794 -1.6148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5975 -1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3870 -1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8134 -0.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0975 -1.0921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1247 -1.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2753 -1.6944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9543 -0.9324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1024 -3.9607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3788 -3.8189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0746 3.1298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6415 4.2822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
28 31 1 0 0 0 0
34 39 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 1 0 0 0 0
16 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.0413 0.7044
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -0.0147 -0.7797
S SKP 5
ID FL5FADGL0003
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c5O)c(c(C2=O)c(c5)OC(=C2OC(C(OC(C4O)OCC(C4O)O)3)OC(CO)C(C3O)O)c(c1)ccc(c1OC)O)O
M END