Mol:FL5FADGA0018
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0996 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0996 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0996 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0996 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3885 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3885 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6774 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6774 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6774 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6774 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3885 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3885 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0338 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0338 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7449 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7449 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7449 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7449 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0338 0.7620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0338 0.7620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0338 -1.5205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0338 -1.5205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4558 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4558 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1804 0.3434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1804 0.3434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9052 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9052 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9052 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9052 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1804 2.0172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1804 2.0172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4558 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4558 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8105 0.7619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8105 0.7619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5594 -0.9064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5594 -0.9064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3885 -1.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3885 -1.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4126 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4126 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9835 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9835 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8088 -2.8980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8088 -2.8980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6389 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6389 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0682 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0682 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2429 -2.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2429 -2.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7626 -2.5015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7626 -2.5015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8149 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8149 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3857 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3857 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2110 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2110 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0413 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0413 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4704 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4704 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6451 -0.6471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6451 -0.6471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8542 -0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8542 -0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9670 -0.2056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9670 -0.2056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6438 -1.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6438 -1.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9755 -1.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9755 -1.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3866 -2.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3866 -2.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2508 -3.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2508 -3.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6389 -3.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6389 -3.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5188 2.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5188 2.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6650 2.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6650 2.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3440 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3440 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1302 1.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1302 1.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3717 1.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3717 1.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6227 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6227 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8506 1.3402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8506 1.3402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6265 2.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6265 2.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9240 3.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9240 3.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7705 2.4119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7705 2.4119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0682 1.1461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0682 1.1461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3395 0.3122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3395 0.3122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1935 2.7110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1935 2.7110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7577 3.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7577 3.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 25 37 1 0 0 0 0 | + | 25 37 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 22 38 1 0 0 0 0 | + | 22 38 1 0 0 0 0 |
| − | 23 39 1 0 0 0 0 | + | 23 39 1 0 0 0 0 |
| − | 24 40 1 0 0 0 0 | + | 24 40 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 41 15 1 0 0 0 0 | + | 41 15 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 18 1 0 0 0 0 | + | 46 18 1 0 0 0 0 |
| − | 32 52 1 0 0 0 0 | + | 32 52 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 16 53 1 0 0 0 0 | + | 16 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 59 -0.0130 -0.6938 | + | M SBV 1 59 -0.0130 -0.6938 |
| − | S SKP 5 | + | S SKP 5 |
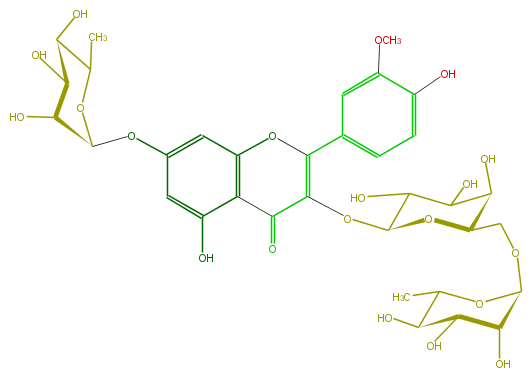
| − | ID FL5FADGA0018 | + | ID FL5FADGA0018 |
| − | FORMULA C34H42O20 | + | FORMULA C34H42O20 |
| − | EXACTMASS 770.226943784 | + | EXACTMASS 770.226943784 |
| − | AVERAGEMASS 770.6852799999999 | + | AVERAGEMASS 770.6852799999999 |
| − | SMILES C(O1)(OC(C(=O)6)=C(Oc(c65)cc(cc5O)OC(C4O)OC(C(O)C4O)C)c(c3)cc(c(c3)O)OC)C(C(O)C(C1COC(C2O)OC(C)C(C2O)O)O)O | + | SMILES C(O1)(OC(C(=O)6)=C(Oc(c65)cc(cc5O)OC(C4O)OC(C(O)C4O)C)c(c3)cc(c(c3)O)OC)C(C(O)C(C1COC(C2O)OC(C)C(C2O)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.0996 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0996 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3885 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6774 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6774 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3885 0.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0338 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7449 -0.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7449 0.3514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0338 0.7620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0338 -1.5205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4558 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1804 0.3434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9052 0.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9052 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1804 2.0172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4558 1.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8105 0.7619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5594 -0.9064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3885 -1.7012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4126 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9835 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8088 -2.8980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6389 -3.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0682 -2.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2429 -2.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7626 -2.5015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8149 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3857 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2110 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0413 -1.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4704 -0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6451 -0.6471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8542 -0.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9670 -0.2056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6438 -1.0115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9755 -1.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3866 -2.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2508 -3.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6389 -3.8580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5188 2.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6650 2.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3440 2.7191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1302 1.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3717 1.1339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6227 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8506 1.3402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6265 2.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9240 3.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7705 2.4119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0682 1.1461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3395 0.3122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1935 2.7110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7577 3.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
31 36 1 0 0 0 0
25 37 1 0 0 0 0
36 37 1 0 0 0 0
22 38 1 0 0 0 0
23 39 1 0 0 0 0
24 40 1 0 0 0 0
29 19 1 0 0 0 0
41 15 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 18 1 0 0 0 0
32 52 1 0 0 0 0
53 54 1 0 0 0 0
16 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 OCH3
M SBV 1 59 -0.0130 -0.6938
S SKP 5
ID FL5FADGA0018
FORMULA C34H42O20
EXACTMASS 770.226943784
AVERAGEMASS 770.6852799999999
SMILES C(O1)(OC(C(=O)6)=C(Oc(c65)cc(cc5O)OC(C4O)OC(C(O)C4O)C)c(c3)cc(c(c3)O)OC)C(C(O)C(C1COC(C2O)OC(C)C(C2O)O)O)O
M END