Mol:FL5FADGA0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.5118 0.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5118 0.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5117 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5117 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7969 -0.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7969 -0.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0821 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0821 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0822 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0822 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7969 1.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7969 1.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6327 -0.6205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6327 -0.6205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3474 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3474 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3475 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3475 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6327 1.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6327 1.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6328 -1.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6328 -1.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2476 1.1892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2476 1.1892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9761 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9761 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7045 1.1893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7045 1.1893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7046 2.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7046 2.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9761 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9761 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2476 2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2476 2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7968 -1.4454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7968 -1.4454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3237 2.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3237 2.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3655 1.1106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3655 1.1106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9180 -0.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9180 -0.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7821 -1.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7821 -1.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0259 -1.4945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0259 -1.4945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6154 -1.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6154 -1.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8657 -2.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8657 -2.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6220 -2.6278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6220 -2.6278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0325 -2.1495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0325 -2.1495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5421 -0.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5421 -0.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5544 -2.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5544 -2.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2243 -2.0455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2243 -2.0455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7137 1.3437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7137 1.3437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0389 0.9541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0389 0.9541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2531 1.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2531 1.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0388 2.4571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0388 2.4571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7136 2.8469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7136 2.8469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4996 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4996 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4185 3.3933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4185 3.3933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9876 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9876 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1878 2.6300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1878 2.6300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3237 1.3133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3237 1.3133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9705 3.0867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9705 3.0867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5376 4.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5376 4.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4376 -3.3216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4376 -3.3216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9076 -4.2397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9076 -4.2397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 25 43 1 0 0 0 0 | + | 25 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 46 0.0057 -0.6357 | + | M SBV 1 46 0.0057 -0.6357 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 48 -0.5719 0.6278 | + | M SBV 2 48 -0.5719 0.6278 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGA0009 | + | ID FL5FADGA0009 |
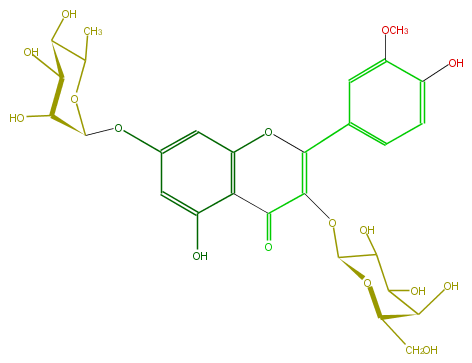
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O | + | SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.5118 0.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5117 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7969 -0.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0821 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0822 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7969 1.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6327 -0.6205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3474 -0.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3475 0.6176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6327 1.0302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6328 -1.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2476 1.1892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9761 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7045 1.1893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7046 2.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9761 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2476 2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7968 -1.4454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3237 2.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3655 1.1106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9180 -0.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7821 -1.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0259 -1.4945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6154 -1.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8657 -2.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6220 -2.6278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0325 -2.1495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5421 -0.9147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5544 -2.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2243 -2.0455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7137 1.3437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0389 0.9541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2531 1.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0388 2.4571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7136 2.8469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4996 2.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4185 3.3933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9876 3.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1878 2.6300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3237 1.3133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9705 3.0867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5376 4.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4376 -3.3216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9076 -4.2397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
21 23 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 20 1 0 0 0 0
41 42 1 0 0 0 0
16 41 1 0 0 0 0
43 44 1 0 0 0 0
25 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 OCH3
M SBV 1 46 0.0057 -0.6357
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 CH2OH
M SBV 2 48 -0.5719 0.6278
S SKP 5
ID FL5FADGA0009
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O
M END