Mol:FL5FACGS0082
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.6573 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6573 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6573 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6573 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3718 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3718 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0863 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0863 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0863 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0863 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3718 3.9458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3718 3.9458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9429 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9429 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2284 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2284 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5139 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5139 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5139 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5139 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2284 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2284 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9429 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9429 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2006 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2006 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9150 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9150 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9150 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9150 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2006 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2006 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2284 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2284 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6295 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6295 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6573 1.0583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6573 1.0583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2006 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2006 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6782 3.8750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6782 3.8750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7148 2.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7148 2.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2316 2.8457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2316 2.8457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8190 2.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8190 2.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0207 2.3401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0207 2.3401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2272 2.1133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2272 2.1133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6397 2.8280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6397 2.8280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4381 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4381 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5951 3.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5951 3.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0750 3.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0750 3.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7148 2.3625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7148 2.3625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2677 2.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2677 2.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1921 1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1921 1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2910 -3.2831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2910 -3.2831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1161 -3.2955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1161 -3.2955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3461 -2.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3461 -2.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9479 -1.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9479 -1.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1227 -1.9257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1227 -1.9257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8926 -2.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8926 -2.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3092 -2.5525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3092 -2.5525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8253 -2.8224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8253 -2.8224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4579 -3.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4579 -3.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6636 -3.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6636 -3.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8075 -2.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8075 -2.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0367 -0.9039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0367 -0.9039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7938 -1.2323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7938 -1.2323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3106 -0.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3106 -0.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0832 -0.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0832 -0.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3261 0.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3261 0.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8092 -0.6135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8092 -0.6135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2999 -0.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2999 -0.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7492 -0.2723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7492 -0.2723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5504 -0.8316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5504 -0.8316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9606 -1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9606 -1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8241 -0.9966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8241 -0.9966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 18 1 0 0 0 0 | + | 26 18 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 48 47 1 1 0 0 0 | + | 48 47 1 1 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 54 1 0 0 0 0 | + | 46 54 1 0 0 0 0 |
| − | 47 55 1 0 0 0 0 | + | 47 55 1 0 0 0 0 |
| − | 48 19 1 0 0 0 0 | + | 48 19 1 0 0 0 0 |
| − | 37 55 1 0 0 0 0 | + | 37 55 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0082 | + | ID FL5FACGS0082 |
| − | KNApSAcK_ID C00013854 | + | KNApSAcK_ID C00013854 |
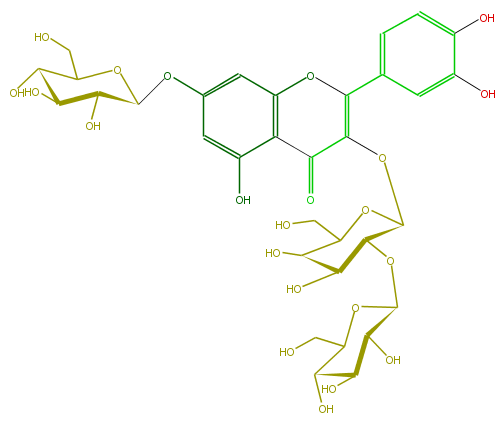
| − | NAME Quercetin 3-glucosyl-(1->2)-galactoside-7-glucoside;2-(3,4-Dihydroxyphenyl)-3-[(2-O-beta-D-glucopyranosyl-beta-D-galactopyranosyl)oxy]-7-(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one | + | NAME Quercetin 3-glucosyl-(1->2)-galactoside-7-glucoside;2-(3,4-Dihydroxyphenyl)-3-[(2-O-beta-D-glucopyranosyl-beta-D-galactopyranosyl)oxy]-7-(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 382143-42-2 | + | CAS_RN 382143-42-2 |
| − | FORMULA C33H40O22 | + | FORMULA C33H40O22 |
| − | EXACTMASS 788.201122964 | + | EXACTMASS 788.201122964 |
| − | AVERAGEMASS 788.6575 | + | AVERAGEMASS 788.6575 |
| − | SMILES c(c6O)cc(cc6O)C(O3)=C(C(=O)c(c(O)4)c3cc(OC(C(O)5)OC(CO)C(O)C5O)c4)OC(O2)C(C(C(O)C2CO)O)OC(C1O)OC(CO)C(C1O)O | + | SMILES c(c6O)cc(cc6O)C(O3)=C(C(=O)c(c(O)4)c3cc(OC(C(O)5)OC(CO)C(O)C5O)c4)OC(O2)C(C(C(O)C2CO)O)OC(C1O)OC(CO)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
2.6573 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6573 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3718 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0863 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0863 3.5333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3718 3.9458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9429 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2284 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5139 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5139 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2284 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9429 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2006 2.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9150 2.2958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9150 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2006 1.0583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2284 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6295 2.7083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6573 1.0583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2006 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6782 3.8750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7148 2.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2316 2.8457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8190 2.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0207 2.3401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2272 2.1133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6397 2.8280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4381 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5951 3.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0750 3.4818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7148 2.3625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2677 2.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1921 1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2910 -3.2831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1161 -3.2955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3461 -2.5029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9479 -1.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1227 -1.9257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8926 -2.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3092 -2.5525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8253 -2.8224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4579 -3.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6636 -3.5478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8075 -2.9784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0367 -0.9039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7938 -1.2323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3106 -0.5889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0832 -0.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3261 0.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8092 -0.6135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2999 -0.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7492 -0.2723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5504 -0.8316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9606 -1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8241 -0.9966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 18 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 44 1 0 0 0 0
45 46 1 1 0 0 0
46 47 1 1 0 0 0
48 47 1 1 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 45 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
45 53 1 0 0 0 0
46 54 1 0 0 0 0
47 55 1 0 0 0 0
48 19 1 0 0 0 0
37 55 1 0 0 0 0
S SKP 8
ID FL5FACGS0082
KNApSAcK_ID C00013854
NAME Quercetin 3-glucosyl-(1->2)-galactoside-7-glucoside;2-(3,4-Dihydroxyphenyl)-3-[(2-O-beta-D-glucopyranosyl-beta-D-galactopyranosyl)oxy]-7-(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one
CAS_RN 382143-42-2
FORMULA C33H40O22
EXACTMASS 788.201122964
AVERAGEMASS 788.6575
SMILES c(c6O)cc(cc6O)C(O3)=C(C(=O)c(c(O)4)c3cc(OC(C(O)5)OC(CO)C(O)C5O)c4)OC(O2)C(C(C(O)C2CO)O)OC(C1O)OC(CO)C(C1O)O
M END