Mol:FL5FACGS0053
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.3581 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3581 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3581 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3581 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8018 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8018 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2455 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2455 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2455 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2455 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8018 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8018 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6892 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6892 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1328 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1328 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1328 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1328 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6892 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6892 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6892 -1.4232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6892 -1.4232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4324 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4324 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8654 0.1586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8654 0.1586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2984 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2984 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2984 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2984 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8654 1.4680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8654 1.4680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4324 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4324 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8018 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8018 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3084 1.4910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3084 1.4910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8530 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8530 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5696 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5696 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8654 2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8654 2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3223 1.6250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3223 1.6250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8969 1.1996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8969 1.1996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4984 1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4984 1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0178 0.8992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0178 0.8992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4433 1.3246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4433 1.3246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8417 1.3147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8417 1.3147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7439 1.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7439 1.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1785 1.6516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1785 1.6516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7977 0.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7977 0.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5506 0.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5506 0.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8904 0.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8904 0.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8904 -0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8904 -0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4019 -0.7376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4019 -0.7376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4656 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4656 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4656 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4656 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9283 -1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9283 -1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3910 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3910 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3910 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3910 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9283 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9283 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8530 -0.5209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8530 -0.5209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8530 -1.5886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8530 -1.5886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9283 -2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9283 -2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0053 | + | ID FL5FACGS0053 |
| − | KNApSAcK_ID C00005970 | + | KNApSAcK_ID C00005970 |
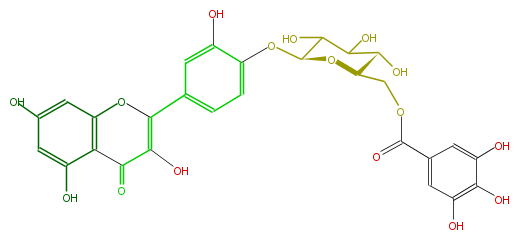
| − | NAME Quercetin 4'-(6''-galloylglucoside) | + | NAME Quercetin 4'-(6''-galloylglucoside) |
| − | CAS_RN 149998-41-4 | + | CAS_RN 149998-41-4 |
| − | FORMULA C28H24O16 | + | FORMULA C28H24O16 |
| − | EXACTMASS 616.1064347199999 | + | EXACTMASS 616.1064347199999 |
| − | AVERAGEMASS 616.48056 | + | AVERAGEMASS 616.48056 |
| − | SMILES c(c5O)c(O)cc(c15)OC(c(c4)cc(O)c(c4)OC(C2O)OC(COC(=O)c(c3)cc(O)c(c(O)3)O)C(C(O)2)O)=C(O)C1=O | + | SMILES c(c5O)c(O)cc(c15)OC(c(c4)cc(O)c(c4)OC(C2O)OC(COC(=O)c(c3)cc(O)c(c(O)3)O)C(C(O)2)O)=C(O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-4.3581 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3581 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8018 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2455 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2455 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8018 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6892 -0.9224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1328 -0.6012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1328 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6892 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6892 -1.4232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4324 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8654 0.1586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2984 0.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2984 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8654 1.4680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4324 1.1407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8018 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3084 1.4910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8530 0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5696 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8654 2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3223 1.6250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8969 1.1996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4984 1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0178 0.8992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4433 1.3246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8417 1.3147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7439 1.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1785 1.6516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7977 0.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5506 0.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8904 0.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8904 -0.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4019 -0.7376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4656 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4656 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9283 -1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3910 -1.3219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3910 -0.7876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9283 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8530 -0.5209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8530 -1.5886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9283 -2.1225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
26 32 1 0 0 0 0
24 19 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
40 42 1 0 0 0 0
39 43 1 0 0 0 0
38 44 1 0 0 0 0
S SKP 8
ID FL5FACGS0053
KNApSAcK_ID C00005970
NAME Quercetin 4'-(6''-galloylglucoside)
CAS_RN 149998-41-4
FORMULA C28H24O16
EXACTMASS 616.1064347199999
AVERAGEMASS 616.48056
SMILES c(c5O)c(O)cc(c15)OC(c(c4)cc(O)c(c4)OC(C2O)OC(COC(=O)c(c3)cc(O)c(c(O)3)O)C(C(O)2)O)=C(O)C1=O
M END