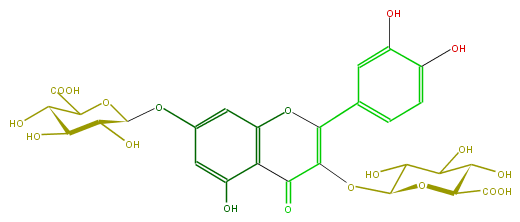
Mol:FL5FACGS0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4381 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4381 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4381 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4381 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7236 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7236 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0090 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0090 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0090 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0090 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7236 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7236 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7055 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7055 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4200 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4200 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4200 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4200 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7055 0.2597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7055 0.2597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7055 -2.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7055 -2.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3198 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3198 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0479 -0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0479 -0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7762 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7762 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7762 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7762 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0479 1.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0479 1.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3198 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3198 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7222 -2.0103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7222 -2.0103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5556 1.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5556 1.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1436 -1.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1436 -1.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2916 0.3400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2916 0.3400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0479 2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0479 2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8504 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8504 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3736 -0.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3736 -0.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6871 -0.0239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6871 -0.0239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0246 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0246 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5060 0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5060 0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1066 0.1477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1066 0.1477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5103 -0.0426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5103 -0.0426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1168 -0.3271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1168 -0.3271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9829 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9829 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4411 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4411 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0547 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0547 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7978 -1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7978 -1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5453 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5453 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9317 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9317 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1887 -1.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1887 -1.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7235 -1.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7235 -1.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7229 -0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7229 -0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6144 -1.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6144 -1.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7253 0.7353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7253 0.7353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7849 0.7353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7849 0.7353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1954 1.6529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1954 1.6529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2551 -1.6029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2551 -1.6029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7849 -0.6852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7849 -0.6852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7849 -2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7849 -2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 28 41 1 0 0 0 0 | + | 28 41 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 35 44 1 0 0 0 0 | + | 35 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 41 42 43 | + | M SAL 1 3 41 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 47 0.6187 -0.5876 | + | M SBV 1 47 0.6187 -0.5876 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 44 45 46 | + | M SAL 2 3 44 45 46 |
| − | M SBL 2 1 50 | + | M SBL 2 1 50 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 50 -0.7097 -0.0811 | + | M SBV 2 50 -0.7097 -0.0811 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGS0037 | + | ID FL5FACGS0037 |
| − | FORMULA C27H26O19 | + | FORMULA C27H26O19 |
| − | EXACTMASS 654.1068286499999 | + | EXACTMASS 654.1068286499999 |
| − | AVERAGEMASS 654.4839400000001 | + | AVERAGEMASS 654.4839400000001 |
| − | SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)1)cc(OC(O2)C(C(C(O)C2C(O)=O)O)O)cc1O | + | SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)1)cc(OC(O2)C(C(C(O)C2C(O)=O)O)O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.4381 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4381 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7236 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0090 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0090 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7236 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7055 -1.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4200 -0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4200 -0.1528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7055 0.2597 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7055 -2.0336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3198 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0479 -0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7762 0.4185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7762 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0479 1.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3198 1.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7222 -2.0103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5556 1.7095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1436 -1.5038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2916 0.3400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0479 2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8504 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3736 -0.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6871 -0.0239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0246 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5060 0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1066 0.1477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5103 -0.0426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1168 -0.3271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9829 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4411 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0547 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7978 -1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5453 -1.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9317 -1.0148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1887 -1.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7235 -1.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7229 -0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6144 -1.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7253 0.7353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7849 0.7353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1954 1.6529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2551 -1.6029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7849 -0.6852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7849 -2.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 21 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 20 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
28 41 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
35 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 41 42 43
M SBL 1 1 47
M SMT 1 ^COOH
M SBV 1 47 0.6187 -0.5876
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 44 45 46
M SBL 2 1 50
M SMT 2 COOH
M SBV 2 50 -0.7097 -0.0811
S SKP 5
ID FL5FACGS0037
FORMULA C27H26O19
EXACTMASS 654.1068286499999
AVERAGEMASS 654.4839400000001
SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)1)cc(OC(O2)C(C(C(O)C2C(O)=O)O)O)cc1O
M END