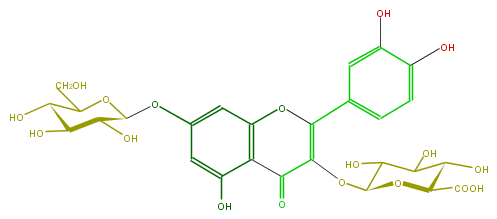
Mol:FL5FACGS0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4409 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4409 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4409 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4409 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7264 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7264 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0119 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0119 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0119 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0119 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7264 0.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7264 0.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7027 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7027 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4172 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4172 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4172 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4172 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7027 0.2777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7027 0.2777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7027 -2.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7027 -2.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3170 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3170 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0451 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0451 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7734 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7734 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7734 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7734 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0451 1.6979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0451 1.6979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3170 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3170 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7264 -2.0453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7264 -2.0453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5528 1.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5528 1.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1407 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1407 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2944 0.3580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2944 0.3580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0451 2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0451 2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1083 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1083 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7219 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7219 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4650 -1.4950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4650 -1.4950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2125 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2125 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5989 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5989 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8558 -1.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8558 -1.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4745 -1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4745 -1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1374 -0.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1374 -0.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3567 -1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3567 -1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8169 0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8169 0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3401 -0.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3401 -0.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6536 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6536 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9912 0.0010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9912 0.0010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4725 0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4725 0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0731 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0731 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5115 0.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5115 0.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0362 -0.3100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0362 -0.3100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9700 -0.4410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9700 -0.4410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8774 -1.6209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8774 -1.6209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9372 -1.6209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9372 -1.6209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3476 -2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3476 -2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7189 0.8273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7189 0.8273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9372 1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9372 1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 37 44 1 0 0 0 0 | + | 37 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 41 42 43 | + | M SAL 1 3 41 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 47 -0.6649 -0.0864 | + | M SBV 1 47 -0.6649 -0.0864 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 49 0.6458 -0.6619 | + | M SBV 2 49 0.6458 -0.6619 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGS0036 | + | ID FL5FACGS0036 |
| − | FORMULA C27H28O18 | + | FORMULA C27H28O18 |
| − | EXACTMASS 640.1275640920001 | + | EXACTMASS 640.1275640920001 |
| − | AVERAGEMASS 640.5004200000001 | + | AVERAGEMASS 640.5004200000001 |
| − | SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)2)cc(cc2O)OC(C(O)1)OC(CO)C(O)C1O | + | SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)2)cc(cc2O)OC(C(O)1)OC(CO)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-1.4409 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4409 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7264 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0119 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0119 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7264 0.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7027 -1.3724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4172 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4172 -0.1348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7027 0.2777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7027 -2.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3170 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0451 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7734 0.4365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7734 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0451 1.6979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3170 1.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7264 -2.0453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5528 1.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1407 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2944 0.3580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0451 2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1083 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7219 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4650 -1.4950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2125 -1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5989 -1.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8558 -1.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4745 -1.0667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1374 -0.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3567 -1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8169 0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3401 -0.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6536 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9912 0.0010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4725 0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0731 0.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5115 0.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0362 -0.3100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9700 -0.4410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8774 -1.6209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9372 -1.6209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3476 -2.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7189 0.8273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9372 1.2341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 20 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 21 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
26 41 1 0 0 0 0
44 45 1 0 0 0 0
37 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 41 42 43
M SBL 1 1 47
M SMT 1 COOH
M SBV 1 47 -0.6649 -0.0864
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 49
M SMT 2 ^ CH2OH
M SBV 2 49 0.6458 -0.6619
S SKP 5
ID FL5FACGS0036
FORMULA C27H28O18
EXACTMASS 640.1275640920001
AVERAGEMASS 640.5004200000001
SMILES C(=C3OC(C5O)OC(C(C5O)O)C(O)=O)(c(c4)ccc(O)c4O)Oc(c(C(=O)3)2)cc(cc2O)OC(C(O)1)OC(CO)C(O)C1O
M END