Mol:FL5FACGL0056
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.1633 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1633 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1633 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1633 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6070 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6070 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0506 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0506 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0506 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0506 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6070 1.3023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6070 1.3023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4943 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4943 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9380 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9380 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9380 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9380 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4943 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4943 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4943 -0.4833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4943 -0.4833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3819 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3819 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8150 0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8150 0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2480 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2480 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2480 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2480 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8150 2.2842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8150 2.2842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3819 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3819 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2899 -0.2115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2899 -0.2115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2445 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2445 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6321 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6321 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1133 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1133 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8633 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8633 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2510 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2510 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5055 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5055 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9716 0.0851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9716 0.0851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7743 0.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7743 0.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8653 0.1153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8653 0.1153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3188 2.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3188 2.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6070 -0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6070 -0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7742 1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7742 1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4922 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4922 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8378 -0.9901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8378 -0.9901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1274 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1274 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7948 -2.2968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7948 -2.2968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8150 2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8150 2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6640 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6640 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0159 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0159 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7197 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7197 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0715 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0715 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7197 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7197 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0159 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0159 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0710 -0.5028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0710 -0.5028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7742 -1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7742 -1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0710 -2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0710 -2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGL0056 | + | ID FL5FACGL0056 |
| − | KNApSAcK_ID C00005958 | + | KNApSAcK_ID C00005958 |
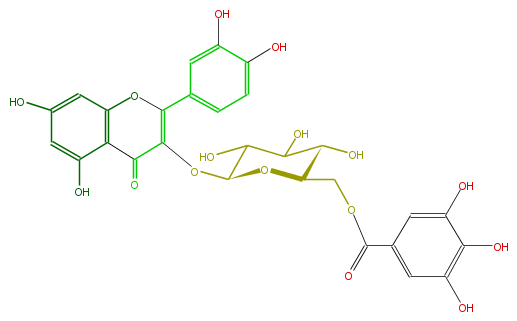
| − | NAME Quercetin 3-(6''-galloylglucoside) | + | NAME Quercetin 3-(6''-galloylglucoside) |
| − | CAS_RN 56316-75-7 | + | CAS_RN 56316-75-7 |
| − | FORMULA C28H24O16 | + | FORMULA C28H24O16 |
| − | EXACTMASS 616.1064347199999 | + | EXACTMASS 616.1064347199999 |
| − | AVERAGEMASS 616.48056 | + | AVERAGEMASS 616.48056 |
| − | SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O | + | SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-4.1633 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1633 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6070 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0506 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0506 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6070 1.3023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4943 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9380 0.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9380 0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4943 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4943 -0.4833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3819 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8150 0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2480 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2480 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8150 2.2842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3819 1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2899 -0.2115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2445 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6321 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1133 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8633 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2510 0.2799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5055 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9716 0.0851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7743 0.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8653 0.1153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3188 2.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6070 -0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7742 1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4922 -0.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8378 -0.9901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1274 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7948 -2.2968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8150 2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6640 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0159 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7197 -2.3302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0715 -1.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7197 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0159 -1.1113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0710 -0.5028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7742 -1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0710 -2.9387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
16 35 1 0 0 0 0
33 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
40 42 1 0 0 0 0
39 43 1 0 0 0 0
38 44 1 0 0 0 0
S SKP 8
ID FL5FACGL0056
KNApSAcK_ID C00005958
NAME Quercetin 3-(6''-galloylglucoside)
CAS_RN 56316-75-7
FORMULA C28H24O16
EXACTMASS 616.1064347199999
AVERAGEMASS 616.48056
SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O
M END