Mol:FL5FACGL0041
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3059 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3059 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3059 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3059 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5948 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5948 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1163 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1163 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1163 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1163 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5948 1.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5948 1.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8274 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8274 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5387 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5387 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5387 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5387 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8274 1.2332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8274 1.2332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8274 -1.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8274 -1.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2494 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2494 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9742 0.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9742 0.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6990 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6990 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6990 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6990 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9742 2.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9742 2.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2494 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2494 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0168 1.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0168 1.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2295 -0.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2295 -0.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5948 -1.2300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5948 -1.2300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8764 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8764 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4473 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4473 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2726 -2.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2726 -2.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1027 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1027 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5320 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5320 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7067 -2.0934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7067 -2.0934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1233 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1233 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2993 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2993 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8702 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8702 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6954 -0.3857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6954 -0.3857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5256 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5256 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9548 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9548 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1296 -0.1140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1296 -0.1140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4418 0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4418 0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5738 0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5738 0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7002 -0.0241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7002 -0.0241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2108 -0.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2108 -0.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4187 -1.2007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4187 -1.2007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8916 -2.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8916 -2.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7971 -3.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7971 -3.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1027 -3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1027 -3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5094 2.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5094 2.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9742 3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9742 3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7617 1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7617 1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2433 0.7035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2433 0.7035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4970 0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4970 0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7192 0.9852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7192 0.9852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3002 1.5249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3002 1.5249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0626 1.2512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0626 1.2512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3962 1.0735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3962 1.0735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9169 0.6768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9169 0.6768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7587 0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7587 0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4785 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4785 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7002 2.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7002 2.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 42 15 1 0 0 0 0 | + | 42 15 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 18 1 0 0 0 0 | + | 47 18 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 59 0.4160 -0.5663 | + | M SBV 1 59 0.4160 -0.5663 |
| − | S SKP 5 | + | S SKP 5 |
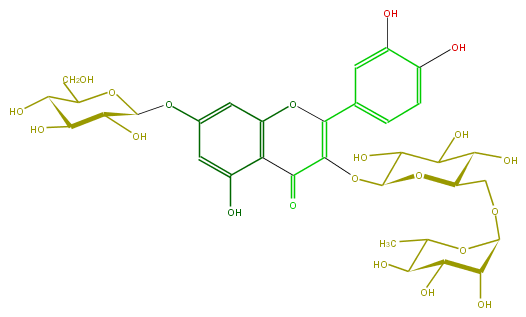
| − | ID FL5FACGL0041 | + | ID FL5FACGL0041 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES c(c5)(c(c(cc5OC(C(O)6)OC(CO)C(O)C6O)1)C(=O)C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)=C(c(c2)ccc(O)c(O)2)O1)O | + | SMILES c(c5)(c(c(cc5OC(C(O)6)OC(CO)C(O)C6O)1)C(=O)C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)=C(c(c2)ccc(O)c(O)2)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.3059 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3059 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5948 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1163 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1163 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5948 1.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8274 -0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5387 0.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5387 0.8226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8274 1.2332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8274 -1.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2494 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9742 0.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6990 1.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6990 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9742 2.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2494 2.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0168 1.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2295 -0.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5948 -1.2300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8764 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4473 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2726 -2.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1027 -2.6009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5320 -1.8576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7067 -2.0934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1233 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2993 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8702 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6954 -0.3857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5256 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9548 0.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1296 -0.1140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4418 0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5738 0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7002 -0.0241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2108 -0.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4187 -1.2007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8916 -2.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7971 -3.0494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1027 -3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5094 2.5378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9742 3.3250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7617 1.3876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2433 0.7035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4970 0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7192 0.9852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3002 1.5249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0626 1.2512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3962 1.0735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9169 0.6768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7587 0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4785 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7002 2.1940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
25 38 1 0 0 0 0
37 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
29 19 1 0 0 0 0
42 15 1 0 0 0 0
16 43 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 44 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 18 1 0 0 0 0
53 54 1 0 0 0 0
49 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 ^ CH2OH
M SBV 1 59 0.4160 -0.5663
S SKP 5
ID FL5FACGL0041
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES c(c5)(c(c(cc5OC(C(O)6)OC(CO)C(O)C6O)1)C(=O)C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)=C(c(c2)ccc(O)c(O)2)O1)O
M END