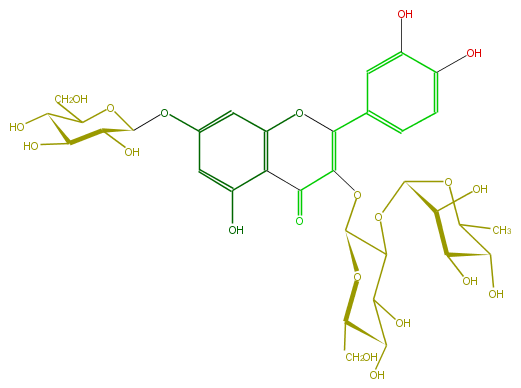
Mol:FL5FACGL0038
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9960 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9960 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9960 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9960 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2816 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2816 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4329 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4329 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4329 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4329 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2816 1.7248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2816 1.7248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1474 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1474 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8618 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8618 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8618 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8618 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1474 1.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1474 1.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1474 -0.5683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1474 -0.5683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5760 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5760 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3042 1.3043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3042 1.3043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0323 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0323 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0323 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0323 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3042 2.9859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3042 2.9859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5760 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5760 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2816 -0.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2816 -0.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7102 1.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7102 1.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7603 2.9857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7603 2.9857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3680 -0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3680 -0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9683 -1.2901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9683 -1.2901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1060 -0.7922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1060 -0.7922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3797 -1.7496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3797 -1.7496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1060 -2.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1060 -2.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9683 -3.2108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9683 -3.2108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6948 -2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6948 -2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8250 -0.4905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8250 -0.4905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2552 -2.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2552 -2.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7024 -3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7024 -3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2599 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2599 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1915 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1915 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5219 -0.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5219 -0.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2930 0.2591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2930 0.2591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3614 0.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3614 0.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0309 -0.3887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0309 -0.3887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8878 0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8878 0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6831 -1.8441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6831 -1.8441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2432 -2.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2432 -2.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2987 -0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2987 -0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3042 3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3042 3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2131 1.6970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2131 1.6970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7364 1.0677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7364 1.0677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0500 1.3347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0500 1.3347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3876 1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3876 1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8689 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8689 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4695 1.5062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4695 1.5062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7644 1.4320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7644 1.4320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4738 1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4738 1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4697 0.8999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4697 0.8999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0853 -3.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0853 -3.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8013 -3.4150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8013 -3.4150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0712 2.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0712 2.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2987 2.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2987 2.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 19 1 0 0 0 0 | + | 45 19 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 51 1 0 0 0 0 | + | 25 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 47 53 1 0 0 0 0 | + | 47 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 57 0.0207 0.7295 | + | M SBV 1 57 0.0207 0.7295 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 59 0.6016 -0.5020 | + | M SBV 2 59 0.6016 -0.5020 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0038 | + | ID FL5FACGL0038 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES OC(C6O)C(C(OC(CO)6)OC(C(=O)4)=C(c(c5)cc(c(c5)O)O)Oc(c42)cc(OC(C(O)3)OC(CO)C(O)C3O)cc2O)OC(C(O)1)OC(C(O)C1O)C | + | SMILES OC(C6O)C(C(OC(CO)6)OC(C(=O)4)=C(c(c5)cc(c(c5)O)O)Oc(c42)cc(OC(C(O)3)OC(CO)C(O)C3O)cc2O)OC(C(O)1)OC(C(O)C1O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-0.9960 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9960 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2816 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4329 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4329 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2816 1.7248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1474 0.0749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8618 0.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8618 1.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1474 1.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1474 -0.5683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5760 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3042 1.3043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0323 1.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0323 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3042 2.9859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5760 2.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2816 -0.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7102 1.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7603 2.9857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3680 -0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9683 -1.2901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1060 -0.7922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3797 -1.7496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1060 -2.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9683 -3.2108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6948 -2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8250 -0.4905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2552 -2.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7024 -3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2599 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1915 -1.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5219 -0.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2930 0.2591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3614 0.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0309 -0.3887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8878 0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6831 -1.8441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2432 -2.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2987 -0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3042 3.8264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2131 1.6970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7364 1.0677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0500 1.3347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3876 1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8689 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4695 1.5062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7644 1.4320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4738 1.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4697 0.8999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0853 -3.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8013 -3.4150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0712 2.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2987 2.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
16 41 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 19 1 0 0 0 0
51 52 1 0 0 0 0
25 51 1 0 0 0 0
53 54 1 0 0 0 0
47 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 ^ CH2OH
M SBV 1 57 0.0207 0.7295
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 ^ CH2OH
M SBV 2 59 0.6016 -0.5020
S SKP 5
ID FL5FACGL0038
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES OC(C6O)C(C(OC(CO)6)OC(C(=O)4)=C(c(c5)cc(c(c5)O)O)Oc(c42)cc(OC(C(O)3)OC(CO)C(O)C3O)cc2O)OC(C(O)1)OC(C(O)C1O)C
M END