Mol:FL5FACGL0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.4612 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4612 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4612 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4612 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7602 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7602 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0592 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0592 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0592 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0592 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7602 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7602 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3582 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3582 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6570 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6570 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6570 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6570 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3582 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3582 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3582 -0.4968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3582 -0.4968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0436 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0436 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7581 1.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7581 1.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4726 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4726 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4726 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4726 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7581 2.9906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7581 2.9906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0436 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0436 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1619 1.7531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1619 1.7531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0678 0.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0678 0.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7602 -0.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7602 -0.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2714 3.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2714 3.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4349 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4349 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2995 -0.6055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2995 -0.6055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0665 -1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0665 -1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2995 -2.2412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2995 -2.2412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4349 -2.6652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4349 -2.6652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2019 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2019 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3917 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3917 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7848 -2.2695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7848 -2.2695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2919 -3.3482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2919 -3.3482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2020 0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2020 0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7595 -0.4752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7595 -0.4752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5623 -0.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5623 -0.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3369 -0.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3369 -0.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7740 0.4084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7740 0.4084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0717 0.0378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0717 0.0378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1406 -0.4583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1406 -0.4583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1818 -0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1818 -0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8462 0.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8462 0.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7581 3.8154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7581 3.8154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0589 -2.8935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0589 -2.8935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5523 -3.4855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5523 -3.4855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1335 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1335 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3419 -3.8154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3419 -3.8154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5931 -2.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5931 -2.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3419 -2.0521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3419 -2.0521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1335 -1.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1335 -1.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8824 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8824 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8922 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8922 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4465 -1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4465 -1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5312 -1.9546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5312 -1.9546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8422 -3.3120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8422 -3.3120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5919 0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5919 0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1619 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1619 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 1 0 0 0 | + | 47 48 1 1 0 0 0 |
| − | 48 43 1 1 0 0 0 | + | 48 43 1 1 0 0 0 |
| − | 47 49 1 0 0 0 0 | + | 47 49 1 0 0 0 0 |
| − | 46 50 1 0 0 0 0 | + | 46 50 1 0 0 0 0 |
| − | 48 51 1 0 0 0 0 | + | 48 51 1 0 0 0 0 |
| − | 43 52 1 0 0 0 0 | + | 43 52 1 0 0 0 0 |
| − | 44 42 1 0 0 0 0 | + | 44 42 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 35 53 1 0 0 0 0 | + | 35 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -0.8179 -0.1164 | + | M SBV 1 59 -0.8179 -0.1164 |
| − | S SKP 5 | + | S SKP 5 |
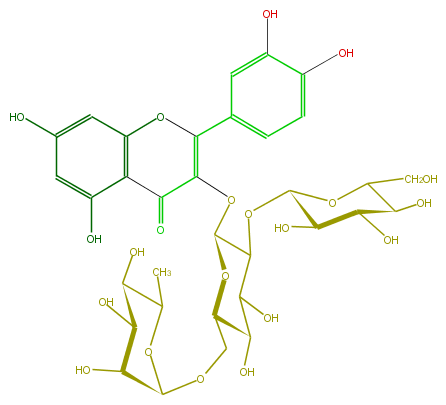
| − | ID FL5FACGL0028 | + | ID FL5FACGL0028 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES C(O)C(O1)C(C(O)C(C(OC(C3OC(=C(c(c6)cc(c(c6)O)O)5)C(=O)c(c(O5)4)c(cc(O)c4)O)C(C(O)C(O3)COC(C(O)2)OC(C)C(O)C(O)2)O)1)O)O | + | SMILES C(O)C(O1)C(C(O)C(C(OC(C3OC(=C(c(c6)cc(c(c6)O)O)5)C(=O)c(c(O5)4)c(cc(O)c4)O)C(C(O)C(O3)COC(C(O)2)OC(C)C(O)C(O)2)O)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-3.4612 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4612 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7602 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0592 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0592 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7602 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3582 0.1343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6570 0.5391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6570 1.3486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3582 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3582 -0.4968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0436 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7581 1.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4726 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4726 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7581 2.9906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0436 2.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1619 1.7531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0678 0.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7602 -0.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2714 3.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4349 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2995 -0.6055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0665 -1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2995 -2.2412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4349 -2.6652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2019 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3917 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7848 -2.2695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2919 -3.3482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2020 0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7595 -0.4752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5623 -0.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3369 -0.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7740 0.4084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0717 0.0378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1406 -0.4583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1818 -0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8462 0.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7581 3.8154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0589 -2.8935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5523 -3.4855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1335 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3419 -3.8154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5931 -2.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3419 -2.0521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1335 -1.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8824 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8922 -0.9394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4465 -1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5312 -1.9546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8422 -3.3120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5919 0.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1619 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
28 31 1 0 0 0 0
34 39 1 0 0 0 0
16 40 1 0 0 0 0
25 41 1 0 0 0 0
41 42 1 0 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 1 0 0 0
48 43 1 1 0 0 0
47 49 1 0 0 0 0
46 50 1 0 0 0 0
48 51 1 0 0 0 0
43 52 1 0 0 0 0
44 42 1 0 0 0 0
53 54 1 0 0 0 0
35 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -0.8179 -0.1164
S SKP 5
ID FL5FACGL0028
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES C(O)C(O1)C(C(O)C(C(OC(C3OC(=C(c(c6)cc(c(c6)O)O)5)C(=O)c(c(O5)4)c(cc(O)c4)O)C(C(O)C(O3)COC(C(O)2)OC(C)C(O)C(O)2)O)1)O)O
M END