Mol:FL5FACGA0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9053 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9053 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9053 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9053 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1499 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1499 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3947 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3947 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3947 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3947 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1499 -1.5106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1499 -1.5106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3607 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3607 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1159 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1159 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1159 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1159 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3607 -1.5106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3607 -1.5106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3620 -4.0454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3620 -4.0454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0670 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0670 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8367 -1.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8367 -1.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6065 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6065 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6065 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6065 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8367 -0.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8367 -0.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0670 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0670 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1499 -4.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1499 -4.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4304 0.0218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4304 0.0218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8807 -3.3748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8807 -3.3748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8367 0.8792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8367 0.8792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9885 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9885 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5800 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5800 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3654 -3.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3654 -3.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1555 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1555 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5641 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5641 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7787 -3.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7787 -3.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3517 -2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3517 -2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1198 -2.6158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1198 -2.6158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5001 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5001 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6862 -1.4958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6862 -1.4958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2250 -1.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2250 -1.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5554 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5554 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7678 -0.8846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7678 -0.8846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5554 -0.1366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5554 -0.1366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2250 0.2500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2250 0.2500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0126 -0.4934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0126 -0.4934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9544 0.8576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9544 0.8576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6349 0.5216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6349 0.5216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6787 -0.0065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6787 -0.0065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9773 -1.2919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9773 -1.2919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0627 3.5394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0627 3.5394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6351 2.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6351 2.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2343 2.2637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2343 2.2637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2343 1.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2343 1.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6618 2.0266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6618 2.0266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0627 2.7300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0627 2.7300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2800 4.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2800 4.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7094 3.7366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7094 3.7366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7094 1.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7094 1.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6605 3.3608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6605 3.3608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9817 4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9817 4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9772 -3.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9772 -3.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9773 -4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9773 -4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 31 1 0 0 0 0 | + | 33 31 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 38 1 0 0 0 0 | + | 45 38 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 57 -0.4022 -0.6308 | + | M SBV 1 57 -0.4022 -0.6308 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 59 -0.8216 -0.1508 | + | M SBV 2 59 -0.8216 -0.1508 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGA0025 | + | ID FL5FACGA0025 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
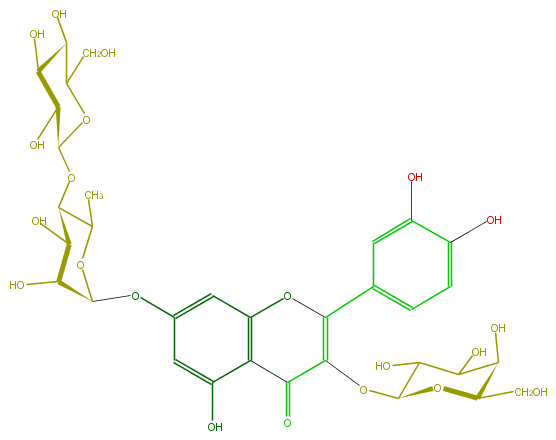
| − | SMILES C(=C5c(c6)cc(c(c6)O)O)(C(=O)c(c4O5)c(cc(c4)OC(C(O)2)OC(C)C(OC(C(O)3)OC(CO)C(O)C3O)C2O)O)OC(O1)C(O)C(C(O)C1CO)O | + | SMILES C(=C5c(c6)cc(c(c6)O)O)(C(=O)c(c4O5)c(cc(c4)OC(C(O)2)OC(C)C(OC(C(O)3)OC(CO)C(O)C3O)C2O)O)OC(O1)C(O)C(C(O)C1CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.9053 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9053 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1499 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3947 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3947 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1499 -1.5106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3607 -3.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1159 -2.8187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1159 -1.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3607 -1.5106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3620 -4.0454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0670 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8367 -1.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6065 -1.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6065 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8367 -0.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0670 -0.4539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1499 -4.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4304 0.0218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8807 -3.3748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8367 0.8792 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9885 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5800 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3654 -3.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1555 -3.5673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5641 -2.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7787 -3.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3517 -2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1198 -2.6158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5001 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6862 -1.4958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2250 -1.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5554 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7678 -0.8846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5554 -0.1366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2250 0.2500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0126 -0.4934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9544 0.8576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6349 0.5216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6787 -0.0065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9773 -1.2919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0627 3.5394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6351 2.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2343 2.2637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2343 1.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6618 2.0266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0627 2.7300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2800 4.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7094 3.7366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7094 1.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6605 3.3608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9817 4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9772 -3.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9773 -4.5366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
1 31 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 31 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 38 1 0 0 0 0
51 52 1 0 0 0 0
47 51 1 0 0 0 0
53 54 1 0 0 0 0
25 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 CH2OH
M SBV 1 57 -0.4022 -0.6308
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 CH2OH
M SBV 2 59 -0.8216 -0.1508
S SKP 5
ID FL5FACGA0025
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES C(=C5c(c6)cc(c(c6)O)O)(C(=O)c(c4O5)c(cc(c4)OC(C(O)2)OC(C)C(OC(C(O)3)OC(CO)C(O)C3O)C2O)O)OC(O1)C(O)C(C(O)C1CO)O
M END