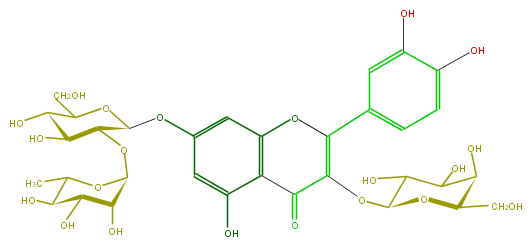
Mol:FL5FACGA0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1250 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1250 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1250 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1250 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3664 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3664 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3922 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3922 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3922 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3922 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3664 0.3657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3664 0.3657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1508 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1508 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9094 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9094 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9094 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9094 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1508 0.3657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1508 0.3657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1508 -2.0692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1508 -2.0692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8647 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8647 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6379 0.0880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6379 0.0880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4111 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4111 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4111 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4111 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6379 1.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6379 1.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8647 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8647 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3664 -2.2619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3664 -2.2619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2385 1.9048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2385 1.9048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6776 -1.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6776 -1.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6379 2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6379 2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7122 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7122 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3018 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3018 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0907 -1.4706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0907 -1.4706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8844 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8844 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2947 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2947 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5058 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5058 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9310 -1.0468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9310 -1.0468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8113 -0.7715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8113 -0.7715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1723 -0.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1723 -0.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9094 0.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9094 0.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4793 0.4764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4793 0.4764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0071 -0.1468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0071 -0.1468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3272 0.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3272 0.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6186 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6186 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1478 0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1478 0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8423 0.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8423 0.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1088 0.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1088 0.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6387 -0.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6387 -0.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7301 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7301 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0178 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0178 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3727 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3727 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6902 -1.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6902 -1.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0034 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0034 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6484 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6484 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3310 -1.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3310 -1.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6597 -1.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6597 -1.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8294 -1.5017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8294 -1.5017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0962 -2.0646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0962 -2.0646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9970 -2.2011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9970 -2.2011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6957 -1.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6957 -1.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6957 -2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6957 -2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3757 0.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3757 0.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6957 1.2306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6957 1.2306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 51 1 0 0 0 0 | + | 25 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 37 53 1 0 0 0 0 | + | 37 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 57 -0.8114 -0.0553 | + | M SBV 1 57 -0.8114 -0.0553 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 59 0.5334 -0.5364 | + | M SBV 2 59 0.5334 -0.5364 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGA0024 | + | ID FL5FACGA0024 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O | + | SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.1250 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1250 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3664 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3922 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3922 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3664 0.3657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1508 -1.3863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9094 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9094 -0.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1508 0.3657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1508 -2.0692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8647 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6379 0.0880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4111 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4111 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6379 1.8734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8647 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3664 -2.2619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2385 1.9048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6776 -1.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6379 2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7122 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3018 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0907 -1.4706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8844 -1.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2947 -0.9856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5058 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9310 -1.0468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8113 -0.7715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1723 -0.3241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9094 0.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4793 0.4764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0071 -0.1468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3272 0.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6186 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1478 0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8423 0.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1088 0.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6387 -0.0809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7301 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0178 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3727 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6902 -1.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0034 -1.6293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6484 -1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3310 -1.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6597 -1.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8294 -1.5017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0962 -2.0646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9970 -2.2011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6957 -1.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6957 -2.7659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3757 0.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6957 1.2306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
1 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 40 1 0 0 0 0
35 31 1 0 0 0 0
51 52 1 0 0 0 0
25 51 1 0 0 0 0
53 54 1 0 0 0 0
37 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 CH2OH
M SBV 1 57 -0.8114 -0.0553
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 ^ CH2OH
M SBV 2 59 0.5334 -0.5364
S SKP 5
ID FL5FACGA0024
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O
M END