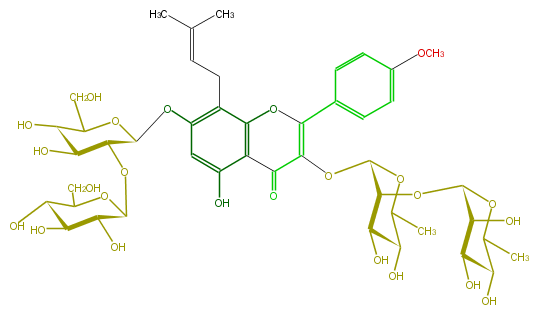
Mol:FL5FABGI0015
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 69 75 0 0 0 0 0 0 0 0999 V2000 | + | 69 75 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4638 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4638 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4638 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4638 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9075 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9075 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3512 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3512 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3512 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3512 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9075 0.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9075 0.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2051 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2051 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7614 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7614 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7614 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7614 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2051 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2051 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2051 -0.7933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2051 -0.7933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4619 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4619 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0288 0.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0288 0.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5958 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5958 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5958 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5958 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0288 2.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0288 2.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4619 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4619 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9075 -0.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9075 -0.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9588 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9588 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3247 -0.3808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3247 -0.3808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9075 1.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9075 1.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2179 0.6176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.2179 0.6176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.7909 0.0540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.7909 0.0540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.1761 0.2931 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.1761 0.2931 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5829 0.2995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5829 0.2995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0140 0.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0140 0.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6420 0.5052 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.6420 0.5052 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.7668 0.6656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7668 0.6656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4519 0.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4519 0.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8239 -0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8239 -0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0765 -1.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0765 -1.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4228 -0.9023 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.4228 -0.9023 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.9959 -1.4659 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.9959 -1.4659 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.3811 -1.2268 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.3811 -1.2268 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7878 -1.2204 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7878 -1.2204 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.2189 -0.7892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2189 -0.7892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8469 -1.0147 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.8469 -1.0147 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.5135 -1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5135 -1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0288 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0288 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4636 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4636 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4636 2.5976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4636 2.5976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0197 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0197 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9075 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9075 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1505 -1.4746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.1505 -1.4746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.7787 -1.8373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7787 -1.8373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5794 -1.1398 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.5794 -1.1398 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.7787 -0.4381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7787 -0.4381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1505 -0.0753 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1505 -0.0753 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3498 -0.7729 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3498 -0.7729 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1201 -0.7729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1201 -0.7729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3119 -2.0768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3119 -2.0768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6272 -2.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6272 -2.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1996 -1.4979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1996 -1.4979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0274 -1.9902 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 4.0274 -1.9902 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.6556 -2.3529 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.6556 -2.3529 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.4563 -1.6554 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 4.4563 -1.6554 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 4.6556 -0.9537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6556 -0.9537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0274 -0.5909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.0274 -0.5909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.2267 -1.2885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.2267 -1.2885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.9970 -1.2885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9970 -1.2885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1887 -2.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1887 -2.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5040 -2.9186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5040 -2.9186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0765 -2.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0765 -2.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2026 2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2026 2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9171 1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9171 1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8772 1.2310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8772 1.2310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8552 1.4397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8552 1.4397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9022 -0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9022 -0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6991 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6991 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 21 40 1 0 0 0 0 | + | 21 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 1 0 0 0 | + | 48 49 1 1 0 0 0 |
| − | 49 44 1 1 0 0 0 | + | 49 44 1 1 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 44 51 1 0 0 0 0 | + | 44 51 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | 46 53 1 0 0 0 0 | + | 46 53 1 0 0 0 0 |
| − | 48 20 1 0 0 0 0 | + | 48 20 1 0 0 0 0 |
| − | 55 54 1 1 0 0 0 | + | 55 54 1 1 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 58 59 1 1 0 0 0 | + | 58 59 1 1 0 0 0 |
| − | 59 54 1 1 0 0 0 | + | 59 54 1 1 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 54 61 1 0 0 0 0 | + | 54 61 1 0 0 0 0 |
| − | 55 62 1 0 0 0 0 | + | 55 62 1 0 0 0 0 |
| − | 56 63 1 0 0 0 0 | + | 56 63 1 0 0 0 0 |
| − | 50 58 1 0 0 0 0 | + | 50 58 1 0 0 0 0 |
| − | 15 64 1 0 0 0 0 | + | 15 64 1 0 0 0 0 |
| − | 64 65 1 0 0 0 0 | + | 64 65 1 0 0 0 0 |
| − | 27 66 1 0 0 0 0 | + | 27 66 1 0 0 0 0 |
| − | 66 67 1 0 0 0 0 | + | 66 67 1 0 0 0 0 |
| − | 37 68 1 0 0 0 0 | + | 37 68 1 0 0 0 0 |
| − | 68 69 1 0 0 0 0 | + | 68 69 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 68 69 | + | M SAL 3 2 68 69 |
| − | M SBL 3 1 74 | + | M SBL 3 1 74 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 74 -3.9022 -0.6163 | + | M SVB 3 74 -3.9022 -0.6163 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 66 67 | + | M SAL 2 2 66 67 |
| − | M SBL 2 1 72 | + | M SBL 2 1 72 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 72 -3.8772 1.231 | + | M SVB 2 72 -3.8772 1.231 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 64 65 | + | M SAL 1 2 64 65 |
| − | M SBL 1 1 70 | + | M SBL 1 1 70 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 70 3.2026 2.1209 | + | M SVB 1 70 3.2026 2.1209 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FABGI0015 | + | ID FL5FABGI0015 |
| − | KNApSAcK_ID C00005832 | + | KNApSAcK_ID C00005832 |
| − | NAME Acuminatoside | + | NAME Acuminatoside |
| − | CAS_RN 142735-71-5 | + | CAS_RN 142735-71-5 |
| − | FORMULA C45H60O24 | + | FORMULA C45H60O24 |
| − | EXACTMASS 984.347452848 | + | EXACTMASS 984.347452848 |
| − | AVERAGEMASS 984.9435 | + | AVERAGEMASS 984.9435 |
| − | SMILES C(=CCc(c5O[C@H](O6)[C@@H](O[C@@H]([C@@H](O)7)OC(CO)[C@@H]([C@H](O)7)O)[C@H]([C@@H](O)C6CO)O)c(c(c(c5)O)1)OC(c(c4)ccc(c4)OC)=C(O[C@@H](O2)[C@@H](O[C@@H](O3)[C@H]([C@H](O)[C@H](O)C3C)O)[C@H](O)[C@@H](C(C)2)O)C(=O)1)(C)C | + | SMILES C(=CCc(c5O[C@H](O6)[C@@H](O[C@@H]([C@@H](O)7)OC(CO)[C@@H]([C@H](O)7)O)[C@H]([C@@H](O)C6CO)O)c(c(c(c5)O)1)OC(c(c4)ccc(c4)OC)=C(O[C@@H](O2)[C@@H](O[C@@H](O3)[C@H]([C@H](O)[C@H](O)C3C)O)[C@H](O)[C@@H](C(C)2)O)C(=O)1)(C)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
69 75 0 0 0 0 0 0 0 0999 V2000
-1.4638 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4638 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9075 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3512 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3512 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9075 0.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2051 -0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7614 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7614 0.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2051 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2051 -0.7933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4619 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0288 0.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5958 1.1159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5958 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0288 2.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4619 1.7706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9075 -0.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9588 0.9922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3247 -0.3808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9075 1.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2179 0.6176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.7909 0.0540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1761 0.2931 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5829 0.2995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0140 0.7307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6420 0.5052 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.7668 0.6656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4519 0.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8239 -0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0765 -1.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4228 -0.9023 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.9959 -1.4659 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3811 -1.2268 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7878 -1.2204 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.2189 -0.7892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8469 -1.0147 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.5135 -1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0288 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4636 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4636 2.5976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0197 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9075 2.9186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1505 -1.4746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.7787 -1.8373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5794 -1.1398 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.7787 -0.4381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1505 -0.0753 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3498 -0.7729 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1201 -0.7729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3119 -2.0768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6272 -2.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1996 -1.4979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0274 -1.9902 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.6556 -2.3529 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.4563 -1.6554 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
4.6556 -0.9537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0274 -0.5909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.2267 -1.2885 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.9970 -1.2885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1887 -2.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5040 -2.9186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0765 -2.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2026 2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9171 1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8772 1.2310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8552 1.4397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9022 -0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6991 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
6 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
31 32 1 0 0 0 0
35 30 1 0 0 0 0
25 19 1 0 0 0 0
21 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
41 43 1 0 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 1 0 0 0
49 44 1 1 0 0 0
49 50 1 0 0 0 0
44 51 1 0 0 0 0
45 52 1 0 0 0 0
46 53 1 0 0 0 0
48 20 1 0 0 0 0
55 54 1 1 0 0 0
55 56 1 0 0 0 0
56 57 1 0 0 0 0
57 58 1 0 0 0 0
58 59 1 1 0 0 0
59 54 1 1 0 0 0
59 60 1 0 0 0 0
54 61 1 0 0 0 0
55 62 1 0 0 0 0
56 63 1 0 0 0 0
50 58 1 0 0 0 0
15 64 1 0 0 0 0
64 65 1 0 0 0 0
27 66 1 0 0 0 0
66 67 1 0 0 0 0
37 68 1 0 0 0 0
68 69 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 68 69
M SBL 3 1 74
M SMT 3 CH2OH
M SVB 3 74 -3.9022 -0.6163
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 66 67
M SBL 2 1 72
M SMT 2 CH2OH
M SVB 2 72 -3.8772 1.231
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 64 65
M SBL 1 1 70
M SMT 1 OCH3
M SVB 1 70 3.2026 2.1209
S SKP 8
ID FL5FABGI0015
KNApSAcK_ID C00005832
NAME Acuminatoside
CAS_RN 142735-71-5
FORMULA C45H60O24
EXACTMASS 984.347452848
AVERAGEMASS 984.9435
SMILES C(=CCc(c5O[C@H](O6)[C@@H](O[C@@H]([C@@H](O)7)OC(CO)[C@@H]([C@H](O)7)O)[C@H]([C@@H](O)C6CO)O)c(c(c(c5)O)1)OC(c(c4)ccc(c4)OC)=C(O[C@@H](O2)[C@@H](O[C@@H](O3)[C@H]([C@H](O)[C@H](O)C3C)O)[C@H](O)[C@@H](C(C)2)O)C(=O)1)(C)C
M END