Mol:FL5FAAGS0060
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8426 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8426 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8426 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8426 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2863 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2863 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2700 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2700 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2700 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2700 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2863 -0.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2863 -0.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8263 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8263 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3826 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3826 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3826 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3826 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8263 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8263 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8263 -1.9335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8263 -1.9335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9387 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9387 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5057 -0.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5057 -0.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0727 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0727 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0727 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0727 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5057 0.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5057 0.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9387 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9387 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2863 -2.0748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2863 -2.0748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8664 0.8859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8664 0.8859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3375 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3375 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3211 -1.4633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3211 -1.4633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3525 -0.3896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3525 -0.3896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9814 -0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9814 -0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4469 -0.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4469 -0.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9311 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9311 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3059 -0.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3059 -0.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7735 -0.5381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7735 -0.5381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8664 -0.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8664 -0.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5600 -0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5600 -0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1406 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1406 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1323 0.0834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1323 0.0834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1323 0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1323 0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7772 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7772 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1033 1.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1033 1.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1172 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1172 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8483 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8483 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2715 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2715 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0005 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0005 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4584 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4584 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1874 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1874 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4584 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4584 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0005 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0005 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3544 1.6054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3544 1.6054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0060 | + | ID FL5FAAGS0060 |
| − | KNApSAcK_ID C00005873 | + | KNApSAcK_ID C00005873 |
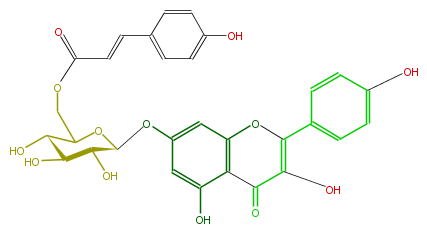
| − | NAME Kaempferol 7-(6''-p-coumarylglucoside);Biondnoid A;Buddlenoid A | + | NAME Kaempferol 7-(6''-p-coumarylglucoside);Biondnoid A;Buddlenoid A |
| − | CAS_RN 142750-32-1 | + | CAS_RN 142750-32-1 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES C(=O)(c32)C(=C(Oc(cc(OC(C(O)5)OC(C(O)C5O)COC(C=Cc(c4)ccc(O)c4)=O)cc3O)2)c(c1)ccc(O)c1)O | + | SMILES C(=O)(c32)C(=C(Oc(cc(OC(C(O)5)OC(C(O)C5O)COC(C=Cc(c4)ccc(O)c4)=O)cc3O)2)c(c1)ccc(O)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.8426 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8426 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2863 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2700 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2700 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2863 -0.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8263 -1.4327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3826 -1.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3826 -0.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8263 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8263 -1.9335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9387 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5057 -0.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0727 -0.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0727 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5057 0.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9387 0.5066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2863 -2.0748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8664 0.8859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3375 -0.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3211 -1.4633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3525 -0.3896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9814 -0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4469 -0.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9311 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3059 -0.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7735 -0.5381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8664 -0.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5600 -0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1406 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1323 0.0834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1323 0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7772 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1033 1.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1172 1.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8483 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2715 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0005 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4584 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1874 1.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4584 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0005 2.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3544 1.6054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 20 1 0 0 0 0
27 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
S SKP 8
ID FL5FAAGS0060
KNApSAcK_ID C00005873
NAME Kaempferol 7-(6''-p-coumarylglucoside);Biondnoid A;Buddlenoid A
CAS_RN 142750-32-1
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES C(=O)(c32)C(=C(Oc(cc(OC(C(O)5)OC(C(O)C5O)COC(C=Cc(c4)ccc(O)c4)=O)cc3O)2)c(c1)ccc(O)c1)O
M END