Mol:FL5FAAGS0051
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7080 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7080 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7080 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7080 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1517 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1517 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5954 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5954 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5954 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5954 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1517 1.9385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1517 1.9385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0391 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0391 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4828 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4828 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4828 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4828 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0391 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0391 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0391 0.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0391 0.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7823 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7823 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2154 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2154 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3516 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3516 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3516 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3516 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2154 3.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2154 3.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7823 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7823 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1517 0.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1517 0.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1453 3.0961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1453 3.0961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2030 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2030 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9195 0.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9195 0.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1349 -0.4871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1349 -0.4871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4933 -0.8499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4933 -0.8499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2940 -0.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2940 -0.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4933 0.5494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4933 0.5494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1349 0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1349 0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0644 0.2146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0644 0.2146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8347 0.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8347 0.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0264 -1.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0264 -1.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4933 -1.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4933 -1.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9142 -0.5104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9142 -0.5104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7099 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7099 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3702 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3702 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4374 -2.5241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4374 -2.5241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7002 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7002 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3588 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3588 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6662 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6662 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2810 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2810 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5883 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5883 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2810 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2810 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6662 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6662 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2030 -2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2030 -2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 32 34 2 0 0 0 0 | + | 32 34 2 0 0 0 0 |
| − | 33 35 2 0 0 0 0 | + | 33 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0051 | + | ID FL5FAAGS0051 |
| − | KNApSAcK_ID C00005864 | + | KNApSAcK_ID C00005864 |
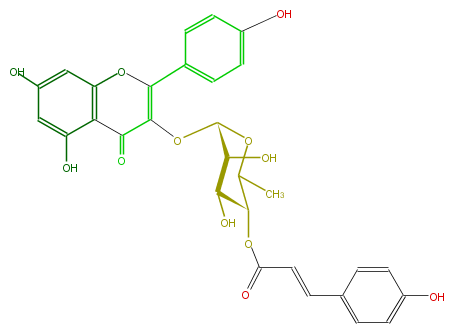
| − | NAME Kaempferol 3-(4''-p-coumarylrhamnoside) | + | NAME Kaempferol 3-(4''-p-coumarylrhamnoside) |
| − | CAS_RN 166321-98-8 | + | CAS_RN 166321-98-8 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES c(c5)(O)cc(O1)c(c(O)5)C(=O)C(OC(C(O)3)OC(C)C(OC(C=Cc(c4)ccc(c4)O)=O)C3O)=C1c(c2)ccc(c2)O | + | SMILES c(c5)(O)cc(O1)c(c(O)5)C(=O)C(OC(C(O)3)OC(C)C(OC(C=Cc(c4)ccc(c4)O)=O)C3O)=C1c(c2)ccc(c2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.7080 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7080 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1517 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5954 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5954 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1517 1.9385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0391 0.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4828 0.9749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4828 1.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0391 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0391 0.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7823 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2154 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3516 2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3516 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2154 3.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7823 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1517 0.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1453 3.0961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2030 1.9385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9195 0.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1349 -0.4871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4933 -0.8499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2940 -0.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4933 0.5494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1349 0.9122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0644 0.2146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8347 0.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0264 -1.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4933 -1.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9142 -0.5104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7099 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3702 -1.9921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4374 -2.5241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7002 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3588 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6662 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2810 -3.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5883 -2.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2810 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6662 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2030 -2.5637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 21 1 0 0 0 0
30 32 1 0 0 0 0
32 33 1 0 0 0 0
32 34 2 0 0 0 0
33 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
S SKP 8
ID FL5FAAGS0051
KNApSAcK_ID C00005864
NAME Kaempferol 3-(4''-p-coumarylrhamnoside)
CAS_RN 166321-98-8
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES c(c5)(O)cc(O1)c(c(O)5)C(=O)C(OC(C(O)3)OC(C)C(OC(C=Cc(c4)ccc(c4)O)=O)C3O)=C1c(c2)ccc(c2)O
M END