Mol:FL5FAAGS0050
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8151 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8151 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8151 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8151 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2588 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2588 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7025 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7025 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7025 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7025 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2588 1.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2588 1.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1462 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1462 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5899 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5899 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5899 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5899 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1462 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1462 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1462 -0.6456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1462 -0.6456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8894 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8894 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3224 0.9363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3224 0.9363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2445 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2445 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2445 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2445 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3224 2.2456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3224 2.2456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8894 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8894 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2588 -0.7869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2588 -0.7869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0382 2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0382 2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3101 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3101 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0266 -0.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0266 -0.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2420 -1.2856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2420 -1.2856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3862 -1.6483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3862 -1.6483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1869 -0.9508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1869 -0.9508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3862 -0.2491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3862 -0.2491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2420 0.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2420 0.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0427 -0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0427 -0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7276 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7276 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0806 -1.8878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0806 -1.8878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3862 -2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3862 -2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8071 -1.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8071 -1.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1809 -0.1306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1809 -0.1306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1809 0.3032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1809 0.3032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7994 -0.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7994 -0.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2654 -0.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2654 -0.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8629 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8629 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8629 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8629 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3454 -1.3992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3454 -1.3992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8278 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8278 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8278 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8278 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3454 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3454 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3101 -1.3991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3101 -1.3991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0050 | + | ID FL5FAAGS0050 |
| − | KNApSAcK_ID C00005863 | + | KNApSAcK_ID C00005863 |
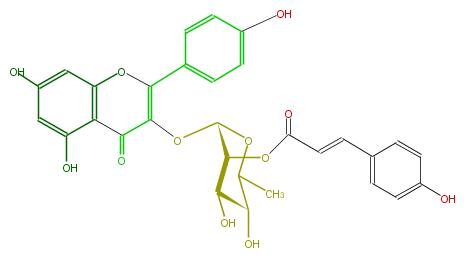
| − | NAME Kaempferol 3-(2-(E)-p-coumarylrhamnoside);3-[[6-Deoxy-2-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-alpha-L-mannopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one | + | NAME Kaempferol 3-(2-(E)-p-coumarylrhamnoside);3-[[6-Deoxy-2-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-alpha-L-mannopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 151455-10-6 | + | CAS_RN 151455-10-6 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES c(C(=C(OC(O5)C(C(O)C(C5C)O)OC(=O)C=Cc(c4)ccc(c4)O)2)Oc(c3)c(c(O)cc3O)C2=O)(c1)ccc(c1)O | + | SMILES c(C(=C(OC(O5)C(C(O)C(C5C)O)OC(=O)C=Cc(c4)ccc(c4)O)2)Oc(c3)c(c(O)cc3O)C2=O)(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.8151 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8151 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2588 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7025 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7025 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2588 1.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1462 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5899 0.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5899 0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1462 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1462 -0.6456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8894 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3224 0.9363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2445 1.2636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2445 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3224 2.2456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8894 1.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2588 -0.7869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0382 2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3101 1.1400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0266 -0.2331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2420 -1.2856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3862 -1.6483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1869 -0.9508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3862 -0.2491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2420 0.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0427 -0.5839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7276 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0806 -1.8878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3862 -2.2976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8071 -1.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1809 -0.1306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1809 0.3032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7994 -0.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2654 -0.2187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8629 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8629 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3454 -1.3992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8278 -1.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8278 -0.5637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3454 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3101 -1.3991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 21 1 0 0 0 0
28 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
S SKP 8
ID FL5FAAGS0050
KNApSAcK_ID C00005863
NAME Kaempferol 3-(2-(E)-p-coumarylrhamnoside);3-[[6-Deoxy-2-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-alpha-L-mannopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 151455-10-6
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES c(C(=C(OC(O5)C(C(O)C(C5C)O)OC(=O)C=Cc(c4)ccc(c4)O)2)Oc(c3)c(c(O)cc3O)C2=O)(c1)ccc(c1)O
M END