Mol:FL5FAAGS0031
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3014 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3014 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3013 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3013 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6003 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6003 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1007 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1007 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1007 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1007 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6004 1.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6004 1.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8017 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8017 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5028 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5028 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5027 1.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5027 1.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8017 1.6892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8017 1.6892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8017 -0.6358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8017 -0.6358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2036 1.6891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2036 1.6891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9181 1.2766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9181 1.2766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6326 1.6890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6326 1.6890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6326 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6326 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9181 2.9266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9181 2.9266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2036 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2036 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6003 -0.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6003 -0.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9538 1.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9538 1.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3469 2.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3469 2.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2331 0.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2331 0.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3359 -0.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3359 -0.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5763 -0.4346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5763 -0.4346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3126 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3126 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8088 -1.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8088 -1.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6352 -1.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6352 -1.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8322 -0.8565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8322 -0.8565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7360 0.2518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7360 0.2518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5909 -0.6816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5909 -0.6816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0104 -1.6153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0104 -1.6153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9359 1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9359 1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4142 0.9274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4142 0.9274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6010 1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6010 1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6947 1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6947 1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3639 1.6700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3639 1.6700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1013 1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1013 1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6154 1.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6154 1.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1807 0.9704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1807 0.9704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8204 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8204 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5019 1.9093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5019 1.9093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4320 2.2979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4320 2.2979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6858 -2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6858 -2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6154 -1.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6154 -1.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5234 -2.9266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5234 -2.9266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.4005 -0.5023 | + | M SBV 1 45 0.4005 -0.5023 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 42 43 44 | + | M SAL 2 3 42 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 48 -0.8770 0.4896 | + | M SBV 2 48 -0.8770 0.4896 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGS0031 | + | ID FL5FAAGS0031 |
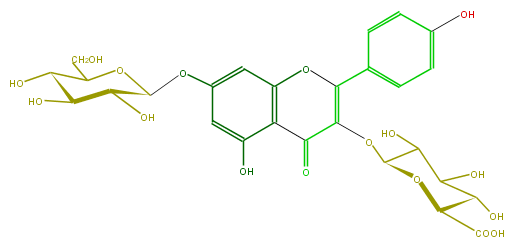
| − | FORMULA C27H28O17 | + | FORMULA C27H28O17 |
| − | EXACTMASS 624.1326494699999 | + | EXACTMASS 624.1326494699999 |
| − | AVERAGEMASS 624.50102 | + | AVERAGEMASS 624.50102 |
| − | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c34)C(C(=C(O3)c(c2)ccc(c2)O)OC(O1)C(C(C(C1C(O)=O)O)O)O)=O)O | + | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c34)C(C(=C(O3)c(c2)ccc(c2)O)OC(O1)C(C(C(C1C(O)=O)O)O)O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.3014 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3013 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6003 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1007 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1007 1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6004 1.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8017 0.0703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5028 0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5027 1.2845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8017 1.6892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8017 -0.6358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2036 1.6891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9181 1.2766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6326 1.6890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6326 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9181 2.9266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2036 2.5141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6003 -0.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9538 1.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3469 2.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2331 0.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3359 -0.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5763 -0.4346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3126 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8088 -1.5157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6352 -1.2149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8322 -0.8565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7360 0.2518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5909 -0.6816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0104 -1.6153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9359 1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4142 0.9274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6010 1.1765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6947 1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3639 1.6700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1013 1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6154 1.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1807 0.9704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8204 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5019 1.9093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4320 2.2979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6858 -2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6154 -1.3555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5234 -2.9266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
25 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.4005 -0.5023
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 42 43 44
M SBL 2 1 48
M SMT 2 COOH
M SBV 2 48 -0.8770 0.4896
S SKP 5
ID FL5FAAGS0031
FORMULA C27H28O17
EXACTMASS 624.1326494699999
AVERAGEMASS 624.50102
SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c34)C(C(=C(O3)c(c2)ccc(c2)O)OC(O1)C(C(C(C1C(O)=O)O)O)O)=O)O
M END