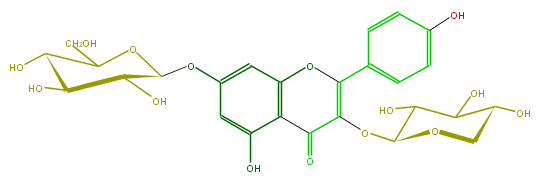
Mol:FL5FAAGS0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1648 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1648 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1648 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1648 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4638 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4638 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2372 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2372 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2372 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2372 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4638 0.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4638 0.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9382 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9382 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6392 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6392 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6392 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6392 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9382 0.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9382 0.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9382 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9382 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3400 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3400 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0545 0.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0545 0.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7690 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7690 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7690 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7690 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0545 1.8327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0545 1.8327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3400 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3400 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4638 -1.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4638 -1.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8655 0.5952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8655 0.5952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3608 1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3608 1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2565 -0.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2565 -0.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4487 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4487 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9602 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9602 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8996 -0.9505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8996 -0.9505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8447 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8447 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3332 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3332 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3938 -0.6413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3938 -0.6413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8485 -0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8485 -0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8245 -0.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8245 -0.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9247 -0.5253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9247 -0.5253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3302 0.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3302 0.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8104 -0.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8104 -0.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4849 0.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4849 0.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5821 0.3352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5821 0.3352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2381 0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2381 0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0567 0.5594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0567 0.5594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9247 0.5671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9247 0.5671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4732 0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4732 0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6999 -0.2252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6999 -0.2252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7759 1.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7759 1.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7801 1.4171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7801 1.4171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.7192 -0.5887 | + | M SBV 1 45 0.7192 -0.5887 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGS0024 | + | ID FL5FAAGS0024 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(O)c(c41)C(=O)C(OC(C3O)OCC(C3O)O)=C(c(c2)ccc(c2)O)O1 | + | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(O)c(c41)C(=O)C(OC(C3O)OCC(C3O)O)=C(c(c2)ccc(c2)O)O1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.1648 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1648 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4638 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2372 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2372 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4638 0.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9382 -1.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6392 -0.6188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6392 0.1907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9382 0.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9382 -1.6547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3400 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0545 0.1827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7690 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7690 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0545 1.8327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3400 1.4202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4638 -1.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8655 0.5952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3608 1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2565 -0.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4487 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9602 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8996 -0.9505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8447 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3332 -0.3729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3938 -0.6413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8485 -0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8245 -0.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9247 -0.5253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3302 0.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8104 -0.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4849 0.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5821 0.3352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2381 0.9914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0567 0.5594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9247 0.5671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4732 0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6999 -0.2252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7759 1.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7801 1.4171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.7192 -0.5887
S SKP 5
ID FL5FAAGS0024
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(O)c(c41)C(=O)C(OC(C3O)OCC(C3O)O)=C(c(c2)ccc(c2)O)O1
M END