Mol:FL5FAAGL0083
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9519 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9519 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9519 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9519 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2375 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2375 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5229 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5229 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5229 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5229 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2375 1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2375 1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1916 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1916 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9060 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9060 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9060 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9060 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1916 1.0483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1916 1.0483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1916 -1.2448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1916 -1.2448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8056 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8056 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5338 0.7867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5338 0.7867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2620 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2620 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2620 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2620 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5338 2.4684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5338 2.4684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8056 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8056 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2375 -1.4263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2375 -1.4263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0413 2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0413 2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6141 1.2186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6141 1.2186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7144 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7144 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9263 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9263 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5399 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5399 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2829 -0.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2829 -0.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0302 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0302 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4167 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4167 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6737 -0.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6737 -0.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2087 -0.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2087 -0.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1254 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1254 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0089 -0.1804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0089 -0.1804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7400 -0.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7400 -0.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1582 -1.2445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1582 -1.2445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1582 -2.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1582 -2.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4556 -2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4556 -2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8085 -2.4526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8085 -2.4526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1485 1.2063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1485 1.2063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6719 0.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6719 0.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9854 0.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9854 0.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3230 0.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3230 0.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8043 1.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8043 1.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4049 1.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4049 1.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8085 0.8252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8085 0.8252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4150 0.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4150 0.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1782 0.4505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1782 0.4505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0183 1.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0183 1.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9950 1.3140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9950 1.3140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 39 20 1 0 0 0 0 | + | 39 20 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 41 45 1 0 0 0 0 | + | 41 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 45 46 | + | M SAL 1 2 45 46 |
| − | M SBL 1 1 50 | + | M SBL 1 1 50 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 50 0.6134 -0.5727 | + | M SBV 1 50 0.6134 -0.5727 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0083 | + | ID FL5FAAGL0083 |
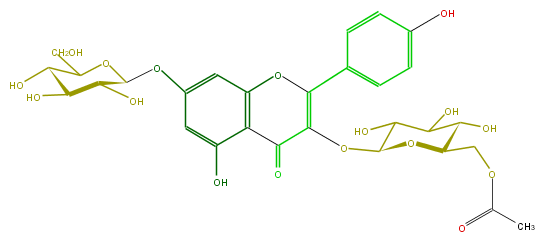
| − | FORMULA C29H32O17 | + | FORMULA C29H32O17 |
| − | EXACTMASS 652.163949598 | + | EXACTMASS 652.163949598 |
| − | AVERAGEMASS 652.55418 | + | AVERAGEMASS 652.55418 |
| − | SMILES C(C1O)(COC(C)=O)OC(OC(C5=O)=C(Oc(c54)cc(cc4O)OC(O3)C(O)C(O)C(O)C(CO)3)c(c2)ccc(c2)O)C(C(O)1)O | + | SMILES C(C1O)(COC(C)=O)OC(OC(C5=O)=C(Oc(c54)cc(cc4O)OC(O3)C(O)C(O)C(O)C(CO)3)c(c2)ccc(c2)O)C(C(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.9519 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9519 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2375 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5229 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5229 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2375 1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1916 -0.6016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9060 -0.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9060 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1916 1.0483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1916 -1.2448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8056 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5338 0.7867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2620 1.2072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2620 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5338 2.4684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8056 2.0479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2375 -1.4263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0413 2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6141 1.2186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7144 -0.6558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9263 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5399 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2829 -0.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0302 -0.7362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4167 -0.0671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6737 -0.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2087 -0.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1254 0.2079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0089 -0.1804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7400 -0.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1582 -1.2445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1582 -2.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4556 -2.4979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8085 -2.4526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1485 1.2063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6719 0.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9854 0.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3230 0.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8043 1.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4049 1.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8085 0.8252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4150 0.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1782 0.4505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0183 1.5882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9950 1.3140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
36 42 1 0 0 0 0
37 43 1 0 0 0 0
38 44 1 0 0 0 0
39 20 1 0 0 0 0
45 46 1 0 0 0 0
41 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 45 46
M SBL 1 1 50
M SMT 1 ^CH2OH
M SBV 1 50 0.6134 -0.5727
S SKP 5
ID FL5FAAGL0083
FORMULA C29H32O17
EXACTMASS 652.163949598
AVERAGEMASS 652.55418
SMILES C(C1O)(COC(C)=O)OC(OC(C5=O)=C(Oc(c54)cc(cc4O)OC(O3)C(O)C(O)C(O)C(CO)3)c(c2)ccc(c2)O)C(C(O)1)O
M END