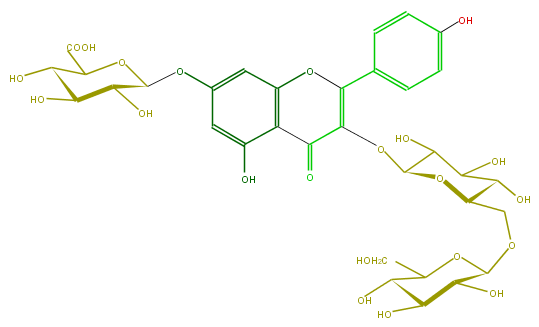
Mol:FL5FAAGL0041
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2696 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2696 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2695 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2695 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5551 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5551 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1594 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1594 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1594 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1594 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5551 2.0478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5551 2.0478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8739 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8739 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5884 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5884 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5884 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5884 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8739 2.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8739 2.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8739 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8739 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3026 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3026 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0307 1.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0307 1.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7590 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7590 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7590 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7590 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0308 3.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0308 3.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3026 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3026 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9839 2.0475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9839 2.0475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5551 -0.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5551 -0.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2467 3.1699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2467 3.1699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6232 0.2617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6232 0.2617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9640 -0.1997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9640 -0.1997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7587 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7587 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3705 -0.8556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3705 -0.8556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0297 -0.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0297 -0.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2352 -0.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2352 -0.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9924 0.5558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9924 0.5558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9676 0.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9676 0.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5160 -0.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5160 -0.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1716 -1.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1716 -1.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6749 -2.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6749 -2.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3919 -3.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3919 -3.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0752 -2.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0752 -2.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8086 -2.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8086 -2.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1309 -2.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1309 -2.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4267 -2.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4267 -2.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9997 -3.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9997 -3.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5953 -3.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5953 -3.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9161 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9161 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3359 -1.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3359 -1.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4401 0.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4401 0.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8121 2.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8121 2.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2638 1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2638 1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4742 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4742 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7123 1.6909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7123 1.6909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2658 2.2447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2658 2.2447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0724 1.9551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0724 1.9551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5160 1.8917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5160 1.8917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0481 1.4354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0481 1.4354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8140 1.1075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8140 1.1075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6656 -2.1218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6656 -2.1218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1083 -0.9647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1083 -0.9647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5417 2.5663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5417 2.5663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4096 3.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4096 3.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5779 3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5779 3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 8 1 0 0 0 0 | + | 41 8 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 18 1 0 0 0 0 | + | 45 18 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 36 51 1 0 0 0 0 | + | 36 51 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 53 55 1 0 0 0 0 | + | 53 55 1 0 0 0 0 |
| − | 47 53 1 0 0 0 0 | + | 47 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 57 0.7611 -0.4032 | + | M SBV 1 57 0.7611 -0.4032 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 53 54 55 | + | M SAL 2 3 53 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 60 0.4693 -0.6111 | + | M SBV 2 60 0.4693 -0.6111 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0041 | + | ID FL5FAAGL0041 |
| − | FORMULA C33H38O22 | + | FORMULA C33H38O22 |
| − | EXACTMASS 786.1854728999999 | + | EXACTMASS 786.1854728999999 |
| − | AVERAGEMASS 786.64162 | + | AVERAGEMASS 786.64162 |
| − | SMILES c(c6)c(ccc6O)C(=C(OC(C(O)4)OC(COC(O5)C(C(O)C(C(CO)5)O)O)C(O)C4O)3)Oc(c(C(=O)3)1)cc(OC(O2)C(C(O)C(O)C(C(O)=O)2)O)cc(O)1 | + | SMILES c(c6)c(ccc6O)C(=C(OC(C(O)4)OC(COC(O5)C(C(O)C(C(CO)5)O)O)C(O)C4O)3)Oc(c(C(=O)3)1)cc(OC(O2)C(C(O)C(O)C(C(O)=O)2)O)cc(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.2696 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2695 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5551 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1594 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1594 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5551 2.0478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8739 0.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5884 0.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5884 1.6353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8739 2.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8739 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3026 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0307 1.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7590 2.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7590 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0308 3.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3026 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9839 2.0475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5551 -0.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2467 3.1699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6232 0.2617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9640 -0.1997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7587 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3705 -0.8556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0297 -0.3942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2352 -0.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9924 0.5558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9676 0.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5160 -0.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1716 -1.0632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6749 -2.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3919 -3.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0752 -2.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8086 -2.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1309 -2.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4267 -2.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9997 -3.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5953 -3.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9161 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3359 -1.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4401 0.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8121 2.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2638 1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4742 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7123 1.6909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2658 2.2447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0724 1.9551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5160 1.8917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0481 1.4354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8140 1.1075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6656 -2.1218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1083 -0.9647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5417 2.5663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4096 3.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5779 3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
22 41 1 0 0 0 0
41 8 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 18 1 0 0 0 0
51 52 1 0 0 0 0
36 51 1 0 0 0 0
53 54 2 0 0 0 0
53 55 1 0 0 0 0
47 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 ^ CH2OH
M SBV 1 57 0.7611 -0.4032
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 53 54 55
M SBL 2 1 60
M SMT 2 ^ COOH
M SBV 2 60 0.4693 -0.6111
S SKP 5
ID FL5FAAGL0041
FORMULA C33H38O22
EXACTMASS 786.1854728999999
AVERAGEMASS 786.64162
SMILES c(c6)c(ccc6O)C(=C(OC(C(O)4)OC(COC(O5)C(C(O)C(C(CO)5)O)O)C(O)C4O)3)Oc(c(C(=O)3)1)cc(OC(O2)C(C(O)C(O)C(C(O)=O)2)O)cc(O)1
M END