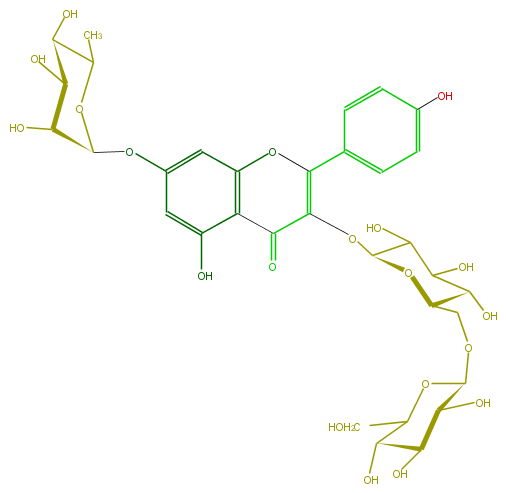
Mol:FL5FAAGL0040
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.7533 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7533 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7533 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7533 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0385 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0385 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3238 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3238 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3239 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3239 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0385 1.9013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0385 1.9013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3909 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3909 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1056 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1056 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1055 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1055 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3908 1.9013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3908 1.9013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3908 -0.3927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3908 -0.3927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8199 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8199 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5484 1.4807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5484 1.4807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2768 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2768 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2768 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2768 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5484 3.1628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5484 3.1628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8199 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8199 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4678 1.9011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4678 1.9011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0384 -0.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0384 -0.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7645 3.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7645 3.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1234 0.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1234 0.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3672 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3672 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1020 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1020 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5564 -1.1905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5564 -1.1905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3127 -0.9153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3127 -0.9153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5777 -0.5872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5777 -0.5872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4800 0.3935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4800 0.3935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1741 -0.4061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1741 -0.4061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6573 -1.3784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6573 -1.3784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0660 -1.3349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0660 -1.3349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4496 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4496 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3493 -4.0608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3493 -4.0608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6906 -3.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6906 -3.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2238 -2.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2238 -2.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4405 -2.7403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4405 -2.7403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0748 -3.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0748 -3.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2813 -4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2813 -4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8699 -4.5404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8699 -4.5404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5299 -3.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5299 -3.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3088 -2.0146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3088 -2.0146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9928 0.1734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9928 0.1734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0194 2.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0194 2.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2124 1.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2124 1.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4684 2.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4684 2.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2124 3.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2124 3.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0194 4.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0194 4.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7633 3.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7633 3.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7425 4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7425 4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3264 4.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3264 4.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3848 3.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3848 3.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6573 2.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6573 2.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1955 -3.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1955 -3.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1339 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1339 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 8 1 0 0 0 0 | + | 41 8 1 0 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 47 42 1 1 0 0 0 | + | 47 42 1 1 0 0 0 |
| − | 46 48 1 0 0 0 0 | + | 46 48 1 0 0 0 0 |
| − | 45 49 1 0 0 0 0 | + | 45 49 1 0 0 0 0 |
| − | 47 50 1 0 0 0 0 | + | 47 50 1 0 0 0 0 |
| − | 42 51 1 0 0 0 0 | + | 42 51 1 0 0 0 0 |
| − | 43 18 1 0 0 0 0 | + | 43 18 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 36 52 1 0 0 0 0 | + | 36 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 58 0.8793 0.0865 | + | M SBV 1 58 0.8793 0.0865 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0040 | + | ID FL5FAAGL0040 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(C1O)(O)C(C(C)OC(Oc(c6)cc(O4)c(c6O)C(C(=C(c(c5)ccc(c5)O)4)OC(C(O)2)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C2O)=O)1)O | + | SMILES C(C1O)(O)C(C(C)OC(Oc(c6)cc(O4)c(c6O)C(C(=C(c(c5)ccc(c5)O)4)OC(C(O)2)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C2O)=O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.7533 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7533 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0385 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3238 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3239 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0385 1.9013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3909 0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1056 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1055 1.4885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3908 1.9013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3908 -0.3927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8199 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5484 1.4807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2768 1.9010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2768 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5484 3.1628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8199 2.7422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4678 1.9011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0384 -0.5740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7645 3.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1234 0.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3672 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1020 -0.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5564 -1.1905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3127 -0.9153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5777 -0.5872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4800 0.3935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1741 -0.4061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6573 -1.3784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0660 -1.3349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4496 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3493 -4.0608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6906 -3.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2238 -2.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4405 -2.7403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0748 -3.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2813 -4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8699 -4.5404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5299 -3.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3088 -2.0146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9928 0.1734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0194 2.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2124 1.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4684 2.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2124 3.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0194 4.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7633 3.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7425 4.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3264 4.2489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3848 3.7639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6573 2.3806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1955 -3.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1339 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
22 41 1 0 0 0 0
41 8 1 0 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 1 0 0 0
47 42 1 1 0 0 0
46 48 1 0 0 0 0
45 49 1 0 0 0 0
47 50 1 0 0 0 0
42 51 1 0 0 0 0
43 18 1 0 0 0 0
52 53 1 0 0 0 0
36 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 ^ CH2OH
M SBV 1 58 0.8793 0.0865
S SKP 5
ID FL5FAAGL0040
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(C1O)(O)C(C(C)OC(Oc(c6)cc(O4)c(c6O)C(C(=C(c(c5)ccc(c5)O)4)OC(C(O)2)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C2O)=O)1)O
M END