Mol:FL5FAAGL0031
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2899 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2899 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2899 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2899 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5889 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5889 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8879 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8879 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8879 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8879 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5889 2.3392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5889 2.3392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1869 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1869 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4859 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4859 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4859 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4859 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1869 2.3392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1869 2.3392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1869 0.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1869 0.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2149 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2149 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9293 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9293 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6438 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6438 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6438 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6438 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9293 3.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9293 3.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2149 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2149 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5889 -0.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5889 -0.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9907 2.3390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9907 2.3390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3581 3.5764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3581 3.5764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3010 0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3010 0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2220 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2220 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1361 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1361 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4791 -0.8897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4791 -0.8897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2545 0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2545 0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3403 0.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3403 0.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9974 -0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9974 -0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3795 -1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3795 -1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1829 -2.2501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1829 -2.2501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8367 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8367 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6934 -2.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6934 -2.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3107 -2.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3107 -2.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8922 -2.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8922 -2.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2425 -3.5103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2425 -3.5103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3068 -3.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3068 -3.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5961 -3.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5961 -3.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0600 -2.4805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0600 -2.4805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8785 -2.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8785 -2.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5222 -2.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5222 -2.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0477 -3.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0477 -3.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4934 -3.5765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4934 -3.5765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6772 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6772 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4689 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4689 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5821 1.0848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5821 1.0848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9418 1.7445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9418 1.7445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1501 1.2875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1501 1.2875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0370 1.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0370 1.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6680 1.8017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6680 1.8017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3343 0.1599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3343 0.1599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9907 0.3411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9907 0.3411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4148 1.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4148 1.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5932 -2.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5932 -2.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3282 -1.5907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3282 -1.5907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 32 31 1 0 0 0 0 | + | 32 31 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 36 32 1 0 0 0 0 | + | 36 32 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 47 42 1 1 0 0 0 | + | 47 42 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 42 49 1 0 0 0 0 | + | 42 49 1 0 0 0 0 |
| − | 43 50 1 0 0 0 0 | + | 43 50 1 0 0 0 0 |
| − | 44 51 1 0 0 0 0 | + | 44 51 1 0 0 0 0 |
| − | 30 46 1 0 0 0 0 | + | 30 46 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 38 52 1 0 0 0 0 | + | 38 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 58 0.7146 -0.5867 | + | M SBV 1 58 0.7146 -0.5867 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0031 | + | ID FL5FAAGL0031 |
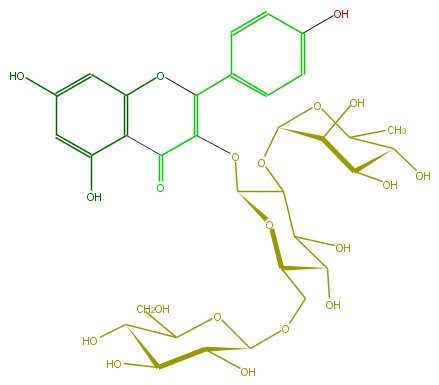
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES OC(C(O)1)C(C(C)OC1OC(C2O)C(OC(C4=O)=C(c(c6)ccc(c6)O)Oc(c5)c4c(O)cc5O)OC(COC(C(O)3)OC(CO)C(O)C3O)C2O)O | + | SMILES OC(C(O)1)C(C(C)OC1OC(C2O)C(OC(C4=O)=C(c(c6)ccc(c6)O)Oc(c5)c4c(O)cc5O)OC(COC(C(O)3)OC(CO)C(O)C3O)C2O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.2899 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2899 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5889 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8879 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8879 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5889 2.3392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1869 0.7203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4859 1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4859 1.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1869 2.3392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1869 0.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2149 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9293 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6438 2.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6438 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9293 3.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2149 3.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5889 -0.0890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9907 2.3390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3581 3.5764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3010 0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2220 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1361 -1.5253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4791 -0.8897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2545 0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3403 0.0018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9974 -0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3795 -1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1829 -2.2501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8367 0.5987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6934 -2.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3107 -2.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8922 -2.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2425 -3.5103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3068 -3.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5961 -3.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0600 -2.4805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8785 -2.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5222 -2.9671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0477 -3.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4934 -3.5765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6772 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4689 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5821 1.0848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9418 1.7445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1501 1.2875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0370 1.0654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6680 1.8017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3343 0.1599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9907 0.3411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4148 1.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5932 -2.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3282 -1.5907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
26 21 1 0 0 0 0
25 30 1 0 0 0 0
22 31 1 0 0 0 0
32 31 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
36 32 1 0 0 0 0
35 41 1 0 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 1 0 0 0
47 42 1 1 0 0 0
47 48 1 0 0 0 0
42 49 1 0 0 0 0
43 50 1 0 0 0 0
44 51 1 0 0 0 0
30 46 1 0 0 0 0
52 53 1 0 0 0 0
38 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 ^CH2OH
M SBV 1 58 0.7146 -0.5867
S SKP 5
ID FL5FAAGL0031
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES OC(C(O)1)C(C(C)OC1OC(C2O)C(OC(C4=O)=C(c(c6)ccc(c6)O)Oc(c5)c4c(O)cc5O)OC(COC(C(O)3)OC(CO)C(O)C3O)C2O)O
M END