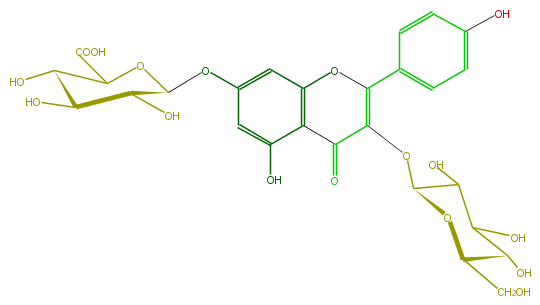
Mol:FL5FAAGL0018
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7035 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7035 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7035 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7035 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0025 0.6023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0025 0.6023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6986 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6986 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6986 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6986 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0025 2.2214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0025 2.2214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3996 0.6024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3996 0.6024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1007 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1007 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1007 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1007 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3996 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3996 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3996 -0.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3996 -0.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8015 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8015 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5160 1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5160 1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2305 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2305 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2306 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2306 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5160 3.4588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5160 3.4588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8015 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8015 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0025 -0.1548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0025 -0.1548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4043 2.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4043 2.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9448 3.4587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9448 3.4587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9435 0.3656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9435 0.3656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0651 -0.2710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0651 -0.2710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0918 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0918 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8505 -0.9717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8505 -0.9717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1727 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1727 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1461 -1.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1461 -1.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3873 -1.1992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3873 -1.1992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5199 0.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5199 0.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2911 -1.4088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2911 -1.4088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4303 -2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4303 -2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7069 2.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7069 2.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2390 1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2390 1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0459 1.7189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0459 1.7189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2334 1.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2334 1.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8239 2.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8239 2.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5607 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5607 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4303 1.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4303 1.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0871 1.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0871 1.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2089 1.2356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2089 1.2356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0139 -2.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0139 -2.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0958 -3.4588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0958 -3.4588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2576 2.6264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2576 2.6264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0680 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0680 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4472 3.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4472 3.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.8412 0.6957 | + | M SBV 1 45 -0.8412 0.6957 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 42 43 44 | + | M SAL 2 3 42 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 48 0.6969 -0.6969 | + | M SBV 2 48 0.6969 -0.6969 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0018 | + | ID FL5FAAGL0018 |
| − | FORMULA C27H28O17 | + | FORMULA C27H28O17 |
| − | EXACTMASS 624.1326494699999 | + | EXACTMASS 624.1326494699999 |
| − | AVERAGEMASS 624.50102 | + | AVERAGEMASS 624.50102 |
| − | SMILES c(c1)c(ccc1C(=C4OC(C(O)5)OC(C(C(O)5)O)CO)Oc(c2C(=O)4)cc(OC(O3)C(C(C(O)C3C(O)=O)O)O)cc2O)O | + | SMILES c(c1)c(ccc1C(=C4OC(C(O)5)OC(C(C(O)5)O)CO)Oc(c2C(=O)4)cc(OC(O3)C(C(C(O)C3C(O)=O)O)O)cc2O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.7035 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7035 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0025 0.6023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6986 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6986 1.8167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0025 2.2214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3996 0.6024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1007 1.0072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1007 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3996 2.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3996 -0.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8015 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5160 1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2305 2.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2306 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5160 3.4588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8015 3.0463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0025 -0.1548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4043 2.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9448 3.4587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9435 0.3656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0651 -0.2710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0918 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8505 -0.9717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1727 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1461 -1.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3873 -1.1992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5199 0.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2911 -1.4088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4303 -2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7069 2.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2390 1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0459 1.7189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2334 1.7277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8239 2.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5607 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4303 1.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0871 1.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2089 1.2356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0139 -2.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0958 -3.4588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2576 2.6264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0680 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4472 3.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
36 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.8412 0.6957
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 42 43 44
M SBL 2 1 48
M SMT 2 ^ COOH
M SBV 2 48 0.6969 -0.6969
S SKP 5
ID FL5FAAGL0018
FORMULA C27H28O17
EXACTMASS 624.1326494699999
AVERAGEMASS 624.50102
SMILES c(c1)c(ccc1C(=C4OC(C(O)5)OC(C(C(O)5)O)CO)Oc(c2C(=O)4)cc(OC(O3)C(C(C(O)C3C(O)=O)O)O)cc2O)O
M END