Mol:FL5FAAGL0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5021 -0.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5021 -0.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2950 -0.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2950 -0.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5719 0.3346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5719 0.3346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0516 0.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0516 0.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7456 0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7456 0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0224 0.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0224 0.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3284 1.7152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3284 1.7152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1919 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1919 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9890 2.1948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9890 2.1948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2658 1.4341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2658 1.4341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9498 1.8250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9498 1.8250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5090 2.8147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5090 2.8147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2269 3.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2269 3.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7571 4.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7571 4.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5696 4.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5696 4.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8519 3.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8519 3.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3215 2.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3215 2.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3686 0.4752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3686 0.4752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7788 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7788 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0998 4.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0998 4.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2935 3.2765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2935 3.2765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6758 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6758 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1873 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1873 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1267 4.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1267 4.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0716 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0716 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5603 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5603 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6209 4.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6209 4.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2095 4.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2095 4.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1533 5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1533 5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0998 4.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0998 4.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1314 -4.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1314 -4.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6487 -4.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6487 -4.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0859 -2.8151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0859 -2.8151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0806 -1.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0806 -1.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6129 -2.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6129 -2.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0453 -3.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0453 -3.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2364 -5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2364 -5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7086 -4.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7086 -4.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4266 -1.9053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4266 -1.9053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8679 4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8679 4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0484 2.9823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0484 2.9823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.7963 -0.1178 | + | M SBV 1 45 -0.7963 -0.1178 |
| − | S SKP 5 | + | S SKP 5 |
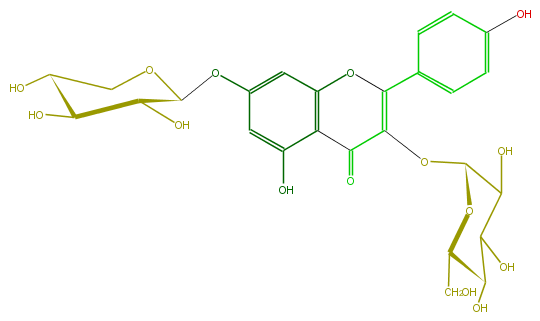
| − | ID FL5FAAGL0013 | + | ID FL5FAAGL0013 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O | + | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.5021 -0.5665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2950 -0.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5719 0.3346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0516 0.9547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7456 0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0224 0.0535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3284 1.7152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1919 2.3353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9890 2.1948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2658 1.4341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9498 1.8250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5090 2.8147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2269 3.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7571 4.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5696 4.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8519 3.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3215 2.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3686 0.4752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7788 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0998 4.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2935 3.2765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6758 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1873 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1267 4.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0716 4.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5603 4.9576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6209 4.6892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2095 4.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1533 5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0998 4.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1314 -4.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6487 -4.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0859 -2.8151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0806 -1.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6129 -2.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0453 -3.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2364 -5.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7086 -4.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4266 -1.9053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8679 4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0484 2.9823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.7963 -0.1178
S SKP 5
ID FL5FAAGL0013
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O
M END