Mol:FL5FAAGL0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3631 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3631 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3631 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3631 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8068 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8068 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2505 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2505 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2505 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2505 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8068 1.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8068 1.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6941 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6941 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1378 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1378 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1378 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1378 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6941 1.7349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6941 1.7349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6941 -0.0507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6941 -0.0507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4182 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4182 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9852 1.4074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9852 1.4074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5522 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5522 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5522 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5522 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9852 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9852 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4182 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4182 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8068 -0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8068 -0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9192 1.7347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9192 1.7347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1190 2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1190 2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8700 -1.1651 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.8700 -1.1651 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.4983 -1.5278 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4983 -1.5278 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2989 -0.8303 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.2989 -0.8303 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4983 -0.1286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4983 -0.1286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8700 0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8700 0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0693 -0.4634 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0693 -0.4634 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8397 -0.4634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8397 -0.4634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0314 -1.7673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0314 -1.7673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3467 -2.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3467 -2.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9192 -1.1884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9192 -1.1884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6959 0.1512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6959 0.1512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1872 -0.8943 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.1872 -0.8943 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.5158 -0.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.5158 -0.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7289 -1.2521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7289 -1.2521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5158 -2.0020 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.5158 -2.0020 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1872 -2.3898 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.1872 -2.3898 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.9743 -1.6443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.9743 -1.6443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9782 -0.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9782 -0.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5059 -1.9512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5059 -1.9512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5230 -3.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5230 -3.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2798 -2.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2798 -2.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7034 -2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7034 -2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 31 1 0 0 0 0 | + | 33 31 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 31 8 1 0 0 0 0 | + | 31 8 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 45 0.2798 -2.2783 | + | M SVB 1 45 0.2798 -2.2783 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGL0009 | + | ID FL5FAAGL0009 |
| − | KNApSAcK_ID C00005167 | + | KNApSAcK_ID C00005167 |
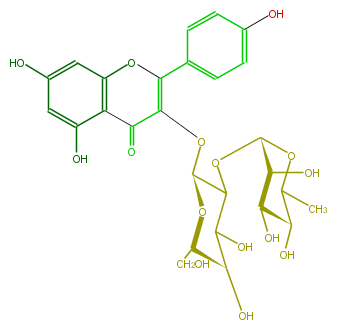
| − | NAME Kaempferol 3-neohesperidoside | + | NAME Kaempferol 3-neohesperidoside |
| − | CAS_RN 32602-81-6 | + | CAS_RN 32602-81-6 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(O)(c5)cc(c(c25)C(C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)=C(O2)c(c1)ccc(c1)O)=O)O | + | SMILES c(O)(c5)cc(c(c25)C(C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)=C(O2)c(c1)ccc(c1)O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.3631 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3631 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8068 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2505 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2505 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8068 1.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6941 0.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1378 0.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1378 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6941 1.7349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6941 -0.0507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4182 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9852 1.4074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5522 1.7347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5522 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9852 2.7168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4182 2.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8068 -0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9192 1.7347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1190 2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8700 -1.1651 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.4983 -1.5278 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2989 -0.8303 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4983 -0.1286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8700 0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0693 -0.4634 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8397 -0.4634 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0314 -1.7673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3467 -2.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9192 -1.1884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6959 0.1512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1872 -0.8943 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.5158 -0.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7289 -1.2521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5158 -2.0020 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1872 -2.3898 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.9743 -1.6443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9782 -0.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5059 -1.9512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5230 -3.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2798 -2.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7034 -2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 31 1 0 0 0 0
25 38 1 0 0 0 0
31 8 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SVB 1 45 0.2798 -2.2783
S SKP 8
ID FL5FAAGL0009
KNApSAcK_ID C00005167
NAME Kaempferol 3-neohesperidoside
CAS_RN 32602-81-6
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(O)(c5)cc(c(c25)C(C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)=C(O2)c(c1)ccc(c1)O)=O)O
M END