Mol:FL5FAAGA0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4922 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4922 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8843 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8843 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8843 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8843 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4922 2.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4922 2.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1001 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1001 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1001 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1001 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2988 0.6809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2988 0.6809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7134 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7134 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7134 1.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7134 1.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2988 2.0330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2988 2.0330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5122 1.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5122 1.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4922 0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4922 0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2988 0.1893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2988 0.1893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0925 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0925 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5095 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5095 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1116 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1116 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1116 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1116 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5095 3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5095 3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0925 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0925 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5347 2.9929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5347 2.9929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7318 -0.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7318 -0.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2773 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2773 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5059 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5059 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8357 0.7045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8357 0.7045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2901 0.1591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2901 0.1591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0615 0.1591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0615 0.1591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1657 0.6020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1657 0.6020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3456 0.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3456 0.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7193 0.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7193 0.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5018 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5018 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4413 -0.0415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4413 -0.0415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0687 -0.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0687 -0.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0687 -1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0687 -1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5122 -1.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5122 -1.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6591 -1.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6591 -1.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6591 -1.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6591 -1.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1253 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1253 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5780 -1.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5780 -1.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0308 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0308 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0308 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0308 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5780 -3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5780 -3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1253 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1253 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5244 -3.0726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5244 -3.0726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 7 13 2 0 0 0 0 | + | 7 13 2 0 0 0 0 |
| − | 9 14 1 0 0 0 0 | + | 9 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 21 26 1 1 0 0 0 | + | 21 26 1 1 0 0 0 |
| − | 8 27 1 0 0 0 0 | + | 8 27 1 0 0 0 0 |
| − | 27 25 1 0 0 0 0 | + | 27 25 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 21 31 1 0 0 0 0 | + | 21 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 2 0 0 0 0 | + | 42 37 2 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0030 | + | ID FL5FAAGA0030 |
| − | KNApSAcK_ID C00005842 | + | KNApSAcK_ID C00005842 |
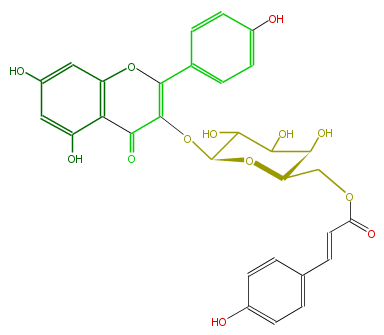
| − | NAME Kaempferol 3-(6''-p-coumarylgalactoside) | + | NAME Kaempferol 3-(6''-p-coumarylgalactoside) |
| − | CAS_RN 72691-81-7,68170-52-5 | + | CAS_RN 72691-81-7,68170-52-5 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES C(c(c5)ccc(O)c5)=CC(=O)OCC(O1)C(O)C(O)C(O)C(OC(=C2c(c4)ccc(c4)O)C(c(c(O)3)c(cc(O)c3)O2)=O)1 | + | SMILES C(c(c5)ccc(O)c5)=CC(=O)OCC(O1)C(O)C(O)C(O)C(OC(=C2c(c4)ccc(c4)O)C(c(c(O)3)c(cc(O)c3)O2)=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.4922 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8843 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8843 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4922 2.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1001 1.7209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1001 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2988 0.6809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7134 1.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7134 1.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2988 2.0330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5122 1.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4922 0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2988 0.1893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0925 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5095 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1116 2.0534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1116 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5095 3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0925 2.7486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5347 2.9929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7318 -0.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2773 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5059 0.3225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8357 0.7045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2901 0.1591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0615 0.1591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1657 0.6020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3456 0.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7193 0.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5018 0.7113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4413 -0.0415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0687 -0.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0687 -1.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5122 -1.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6591 -1.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6591 -1.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1253 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5780 -1.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0308 -2.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0308 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5780 -3.0962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1253 -2.7803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5244 -3.0726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
5 11 1 0 0 0 0
1 12 1 0 0 0 0
7 13 2 0 0 0 0
9 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
17 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
21 26 1 1 0 0 0
8 27 1 0 0 0 0
27 25 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
22 30 1 0 0 0 0
21 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
42 37 2 0 0 0 0
40 43 1 0 0 0 0
S SKP 8
ID FL5FAAGA0030
KNApSAcK_ID C00005842
NAME Kaempferol 3-(6''-p-coumarylgalactoside)
CAS_RN 72691-81-7,68170-52-5
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES C(c(c5)ccc(O)c5)=CC(=O)OCC(O1)C(O)C(O)C(O)C(OC(=C2c(c4)ccc(c4)O)C(c(c(O)3)c(cc(O)c3)O2)=O)1
M END