Mol:FL5FAAGA0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1355 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1355 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1355 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1355 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5792 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5792 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0229 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0229 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0229 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0229 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5792 1.5906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5792 1.5906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4666 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4666 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9103 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9103 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9103 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9103 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4666 1.5906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4666 1.5906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4666 -0.1950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4666 -0.1950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3542 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3542 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2128 1.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2128 1.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7798 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7798 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7798 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7798 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2128 2.5725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2128 2.5725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3542 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3542 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5792 -0.3363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5792 -0.3363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6916 1.5905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6916 1.5905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3466 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3466 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4699 -1.9900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4699 -1.9900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1583 -2.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1583 -2.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0410 -1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0410 -1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1583 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1583 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4699 -0.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4699 -0.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2706 -1.2883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2706 -1.2883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4699 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4699 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0035 -0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0035 -0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4209 -1.9219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4209 -1.9219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0101 0.0938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0101 0.0938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5257 -0.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5257 -0.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2682 -0.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2682 -0.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9847 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9847 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4641 0.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4641 0.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8145 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8145 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7219 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7219 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6937 -1.0123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6937 -1.0123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2978 -0.1096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2978 -0.1096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9693 -2.1354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9693 -2.1354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9771 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9771 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6916 -0.1552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6916 -0.1552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8899 -2.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8899 -2.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1755 -2.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1755 -2.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 21 42 1 0 0 0 0 | + | 21 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 -5.7327 4.2251 | + | M SBV 1 44 -5.7327 4.2251 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -6.6658 4.0282 | + | M SBV 2 46 -6.6658 4.0282 |
| − | S SKP 8 | + | S SKP 8 |
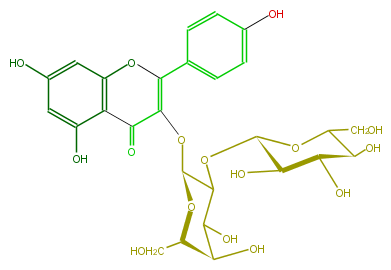
| − | ID FL5FAAGA0005 | + | ID FL5FAAGA0005 |
| − | KNApSAcK_ID C00005157 | + | KNApSAcK_ID C00005157 |
| − | NAME Panasenoside | + | NAME Panasenoside |
| − | CAS_RN 31512-06-8 | + | CAS_RN 31512-06-8 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O | + | SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.1355 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1355 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5792 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0229 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0229 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5792 1.5906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4666 0.3059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9103 0.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9103 1.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4666 1.5906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4666 -0.1950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3542 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2128 1.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7798 1.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7798 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2128 2.5725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3542 2.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5792 -0.3363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6916 1.5905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3466 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4699 -1.9900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1583 -2.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0410 -1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1583 -0.9535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4699 -0.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2706 -1.2883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4699 0.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0035 -0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4209 -1.9219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0101 0.0938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5257 -0.5868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2682 -0.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9847 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4641 0.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8145 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7219 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6937 -1.0123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2978 -0.1096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9693 -2.1354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9771 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6916 -0.1552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8899 -2.1600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1755 -2.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
30 28 1 0 0 0 0
33 38 1 0 0 0 0
22 39 1 0 0 0 0
27 8 1 0 0 0 0
34 40 1 0 0 0 0
40 41 1 0 0 0 0
21 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 -5.7327 4.2251
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -6.6658 4.0282
S SKP 8
ID FL5FAAGA0005
KNApSAcK_ID C00005157
NAME Panasenoside
CAS_RN 31512-06-8
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(O2)(c(C(C(OC(C4OC(C5O)OC(C(C5O)O)CO)OC(CO)C(C(O)4)O)=C2c(c3)ccc(c3)O)=O)1)cc(O)cc1O
M END