Mol:FL5FA9GS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9520 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9520 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9520 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9520 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3957 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3957 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8394 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8394 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8394 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8394 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3957 1.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3957 1.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2831 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2831 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7268 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7268 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7268 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7268 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2831 1.5286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2831 1.5286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2831 -0.2570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2831 -0.2570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1707 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1707 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6037 1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6037 1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0367 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0367 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0367 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0367 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6037 2.5105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6037 2.5105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1707 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1707 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9962 0.0972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9962 0.0972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5081 1.5284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5081 1.5284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3957 -0.3983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3957 -0.3983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0336 -1.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0336 -1.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8536 -1.3708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8536 -1.3708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4625 -0.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4625 -0.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6618 0.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6618 0.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0336 0.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0336 0.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2329 -0.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2329 -0.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0032 -0.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0032 -0.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0336 -1.7257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0336 -1.7257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9155 -1.9895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9155 -1.9895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0827 -1.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0827 -1.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1736 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1736 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7860 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7860 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5315 -1.9116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5315 -1.9116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2814 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2814 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6692 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6692 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9236 -1.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9236 -1.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4537 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4537 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2306 -1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2306 -1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4686 -0.7048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4686 -0.7048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7936 -2.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7936 -2.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5081 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5081 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 29 32 1 0 0 0 0 | + | 29 32 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 -5.8694 8.5766 | + | M SBV 1 44 -5.8694 8.5766 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FA9GS0003 | + | ID FL5FA9GS0003 |
| − | KNApSAcK_ID C00005124 | + | KNApSAcK_ID C00005124 |
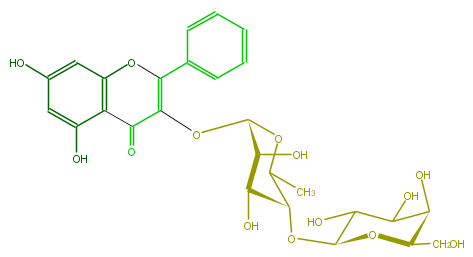
| − | NAME Galangin 3-galactosyl-(1->4)-rhamnoside | + | NAME Galangin 3-galactosyl-(1->4)-rhamnoside |
| − | CAS_RN 73030-72-5 | + | CAS_RN 73030-72-5 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
| − | SMILES OC(C5O)C(O)C(OC5CO)OC(C(O)1)C(C)OC(OC(=C(c(c4)cccc4)3)C(=O)c(c(O)2)c(O3)cc(O)c2)C(O)1 | + | SMILES OC(C5O)C(O)C(OC5CO)OC(C(O)1)C(C)OC(OC(=C(c(c4)cccc4)3)C(=O)c(c(O)2)c(O3)cc(O)c2)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-3.9520 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9520 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3957 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8394 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8394 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3957 1.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2831 0.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7268 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7268 1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2831 1.5286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2831 -0.2570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1707 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6037 1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0367 1.5284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0367 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6037 2.5105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1707 2.1831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9962 0.0972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5081 1.5284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3957 -0.3983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0336 -1.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8536 -1.3708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4625 -0.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6618 0.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0336 0.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2329 -0.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0032 -0.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0336 -1.7257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9155 -1.9895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0827 -1.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1736 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7860 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5315 -1.9116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2814 -2.1247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6692 -1.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9236 -1.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4537 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2306 -1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4686 -0.7048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7936 -2.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5081 -2.5105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 8 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
25 18 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
29 32 1 0 0 0 0
34 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 -5.8694 8.5766
S SKP 8
ID FL5FA9GS0003
KNApSAcK_ID C00005124
NAME Galangin 3-galactosyl-(1->4)-rhamnoside
CAS_RN 73030-72-5
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES OC(C5O)C(O)C(OC5CO)OC(C(O)1)C(C)OC(OC(=C(c(c4)cccc4)3)C(=O)c(c(O)2)c(O3)cc(O)c2)C(O)1
M END