Mol:FL5F1LGL0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3246 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3246 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3246 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3246 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7683 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7683 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2120 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2120 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2120 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2120 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7683 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7683 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6557 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6557 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0994 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0994 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0994 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0994 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6557 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6557 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6557 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6557 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4567 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4567 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0237 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0237 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5906 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5906 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5906 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5906 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0237 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0237 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4567 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4567 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8807 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8807 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1574 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1574 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2947 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2947 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0833 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0833 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4119 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4119 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.6250 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6250 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4119 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4119 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0833 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.0833 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8704 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.8704 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.8904 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8904 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4020 -1.8751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4020 -1.8751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4191 -3.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4191 -3.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9605 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.9605 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.6859 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6859 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1645 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1645 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9863 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9863 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2609 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2609 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7822 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.7822 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4494 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4494 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4014 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4014 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8375 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8375 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8807 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8807 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0237 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0237 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1101 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1101 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1064 -2.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1064 -2.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1062 -2.7599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1062 -2.7599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 34 27 1 0 0 0 0 | + | 34 27 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 17 41 1 0 0 0 0 | + | 17 41 1 0 0 0 0 |
| − | 24 42 1 0 0 0 0 | + | 24 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 46 0.886 -2.6145 | + | M SVB 1 46 0.886 -2.6145 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5F1LGL0001 | + | ID FL5F1LGL0001 |
| − | KNApSAcK_ID C00005627 | + | KNApSAcK_ID C00005627 |
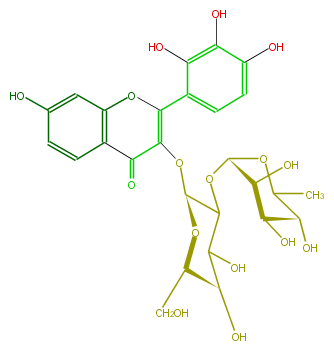
| − | NAME 3,7,2',3',4'-Pentahydroxyflavone 3-neohesperidoside | + | NAME 3,7,2',3',4'-Pentahydroxyflavone 3-neohesperidoside |
| − | CAS_RN 83841-48-9 | + | CAS_RN 83841-48-9 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES O(C=2c(c5)c(c(c(c5)O)O)O)c(c1C(=O)C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)2)cc(O)cc1 | + | SMILES O(C=2c(c5)c(c(c(c5)O)O)O)c(c1C(=O)C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)2)cc(O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.3246 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3246 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7683 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2120 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2120 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7683 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6557 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0994 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0994 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6557 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6557 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4567 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0237 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5906 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5906 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0237 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4567 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8807 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1574 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2947 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0833 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4119 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6250 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4119 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0833 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8704 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.8904 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4020 -1.8751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4191 -3.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9605 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.6859 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1645 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9863 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2609 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7822 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4494 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4014 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8375 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8807 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0237 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1101 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1064 -2.7812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1062 -2.7599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
34 27 1 0 0 0 0
16 40 1 0 0 0 0
17 41 1 0 0 0 0
24 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SVB 1 46 0.886 -2.6145
S SKP 8
ID FL5F1LGL0001
KNApSAcK_ID C00005627
NAME 3,7,2',3',4'-Pentahydroxyflavone 3-neohesperidoside
CAS_RN 83841-48-9
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES O(C=2c(c5)c(c(c(c5)O)O)O)c(c1C(=O)C(O[C@@H](C3O[C@H]([C@H]4O)OC(C)[C@H]([C@H]4O)O)O[C@H](CO)[C@@H](C(O)3)O)2)cc(O)cc1
M END