Mol:FL5F1GNS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 26 29 0 0 0 0 0 0 0 0999 V2000 | + | 26 29 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.2827 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2827 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2827 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2827 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7264 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7264 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1701 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1701 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1701 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1701 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7264 0.1138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7264 0.1138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6138 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6138 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0575 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0575 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0575 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0575 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6138 0.1138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6138 0.1138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6138 -1.6718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6138 -1.6718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4986 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4986 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0655 -0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0655 -0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6325 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6325 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6325 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6325 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0655 1.0957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0655 1.0957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4986 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4986 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2552 -0.0887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2552 -0.0887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6400 0.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6400 0.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2552 0.9706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2552 0.9706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8085 -1.3498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8085 -1.3498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6745 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6745 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8063 1.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8063 1.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5816 1.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5816 1.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6400 0.4113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6400 0.4113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1401 1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1401 1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 16 23 1 0 0 0 0 | + | 16 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 1 25 1 0 0 0 0 | + | 1 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 25 26 | + | M SAL 3 2 25 26 |
| − | M SBL 3 1 28 | + | M SBL 3 1 28 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 28 -2.64 0.4113 | + | M SVB 3 28 -2.64 0.4113 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 23 24 | + | M SAL 2 2 23 24 |
| − | M SBL 2 1 26 | + | M SBL 2 1 26 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 26 1.3489 1.6718 | + | M SVB 2 26 1.3489 1.6718 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 21 22 | + | M SAL 1 2 21 22 |
| − | M SBL 1 1 24 | + | M SBL 1 1 24 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 24 0.1527 -1.0348 | + | M SVB 1 24 0.1527 -1.0348 |
| − | S SKP 8 | + | S SKP 8 |
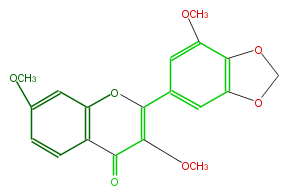
| − | ID FL5F1GNS0002 | + | ID FL5F1GNS0002 |
| − | KNApSAcK_ID C00005049 | + | KNApSAcK_ID C00005049 |
| − | NAME Kanugin | + | NAME Kanugin |
| − | CAS_RN 574-03-8 | + | CAS_RN 574-03-8 |
| − | FORMULA C19H16O7 | + | FORMULA C19H16O7 |
| − | EXACTMASS 356.089602866 | + | EXACTMASS 356.089602866 |
| − | AVERAGEMASS 356.32614 | + | AVERAGEMASS 356.32614 |
| − | SMILES c(c12)c(C(O4)=C(C(c(c43)ccc(OC)c3)=O)OC)cc(c1OCO2)OC | + | SMILES c(c12)c(C(O4)=C(C(c(c43)ccc(OC)c3)=O)OC)cc(c1OCO2)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
26 29 0 0 0 0 0 0 0 0999 V2000
-2.2827 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2827 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7264 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1701 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1701 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7264 0.1138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6138 -1.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0575 -0.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0575 -0.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6138 0.1138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6138 -1.6718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4986 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0655 -0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6325 0.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6325 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0655 1.0957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4986 0.7683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2552 -0.0887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6400 0.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2552 0.9706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8085 -1.3498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6745 -1.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8063 1.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5816 1.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6400 0.4113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1401 1.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
14 18 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 0 0 0 0
20 15 1 0 0 0 0
8 21 1 0 0 0 0
21 22 1 0 0 0 0
16 23 1 0 0 0 0
23 24 1 0 0 0 0
1 25 1 0 0 0 0
25 26 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 25 26
M SBL 3 1 28
M SMT 3 OCH3
M SVB 3 28 -2.64 0.4113
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 23 24
M SBL 2 1 26
M SMT 2 OCH3
M SVB 2 26 1.3489 1.6718
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 21 22
M SBL 1 1 24
M SMT 1 OCH3
M SVB 1 24 0.1527 -1.0348
S SKP 8
ID FL5F1GNS0002
KNApSAcK_ID C00005049
NAME Kanugin
CAS_RN 574-03-8
FORMULA C19H16O7
EXACTMASS 356.089602866
AVERAGEMASS 356.32614
SMILES c(c12)c(C(O4)=C(C(c(c43)ccc(OC)c3)=O)OC)cc(c1OCO2)OC
M END