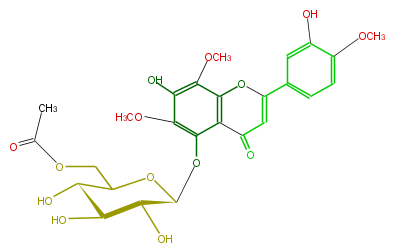
Mol:FL3FGCGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 43 0 0 0 0 0 0 0 0999 V2000 | + | 40 43 0 0 0 0 0 0 0 0999 V2000 |
| − | -6.5233 1.6379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5233 1.6379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0628 1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0628 1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1082 2.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1082 2.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5803 2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5803 2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0069 2.4005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0069 2.4005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.9614 1.8816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.9614 1.8816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6257 3.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6257 3.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0977 3.4382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0977 3.4382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.5243 3.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.5243 3.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.4789 2.6207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.4789 2.6207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4228 3.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4228 3.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.9962 3.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.9962 3.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.0425 3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.0425 3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.5236 4.1127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.5236 4.1127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.9584 3.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.9584 3.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.9122 3.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.9122 3.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.4310 3.0550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.4310 3.0550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6824 2.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6824 2.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5084 1.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5084 1.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -9.3886 2.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -9.3886 2.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9638 1.0736 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.9638 1.0736 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.7020 1.8865 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.7020 1.8865 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -4.3644 2.3290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -4.3644 2.3290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -4.7816 2.9115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -4.7816 2.9115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -4.9096 2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9096 2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2514 1.7212 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -4.2514 1.7212 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.3604 0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3604 0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2088 1.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2088 1.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0234 3.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0234 3.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4389 1.1619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4389 1.1619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0357 0.5860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0357 0.5860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1628 -0.1349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1628 -0.1349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8064 -0.3692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8064 -0.3692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8446 -0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8446 -0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7612 1.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7612 1.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3459 0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3459 0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.5616 1.7672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.5616 1.7672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -8.5439 1.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -8.5439 1.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -9.6504 4.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -9.6504 4.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -9.7223 4.8693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -9.7223 4.8693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 16 1 0 0 0 0 | + | 20 16 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 32 34 2 0 0 0 0 | + | 32 34 2 0 0 0 0 |
| − | 2 35 1 0 0 0 0 | + | 2 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 6 37 1 0 0 0 0 | + | 6 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 42 | + | M SBL 3 1 42 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 42 3.1301 1.8019 | + | M SVB 3 42 3.1301 1.8019 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 37 38 | + | M SAL 2 2 37 38 |
| − | M SBL 2 1 40 | + | M SBL 2 1 40 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 40 0.0642 1.3987 | + | M SVB 2 40 0.0642 1.3987 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 38 -1.3518 0.1923 | + | M SVB 1 38 -1.3518 0.1923 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FGCGS0004 | + | ID FL3FGCGS0004 |
| − | KNApSAcK_ID C00004440 | + | KNApSAcK_ID C00004440 |
| − | NAME 5,7,3'-Trihydroxy-6.8,4'-trimethoxyflavone 5-(6''-acetylglucoside) | + | NAME 5,7,3'-Trihydroxy-6.8,4'-trimethoxyflavone 5-(6''-acetylglucoside) |
| − | CAS_RN 72209-43-9 | + | CAS_RN 72209-43-9 |
| − | FORMULA C26H28O14 | + | FORMULA C26H28O14 |
| − | EXACTMASS 564.147905604 | + | EXACTMASS 564.147905604 |
| − | AVERAGEMASS 564.49212 | + | AVERAGEMASS 564.49212 |
| − | SMILES O=C(c23)C=C(Oc2c(c(O)c(OC)c3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4COC(C)=O)O)OC)c(c1)ccc(c1O)OC | + | SMILES O=C(c23)C=C(Oc2c(c(O)c(OC)c3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4COC(C)=O)O)OC)c(c1)ccc(c1O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/ 40 43 0 0 0 0 0 0 0 0999 V2000 -6.5233 1.6379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.0628 1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.1082 2.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.5803 2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -7.0069 2.4005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.9614 1.8816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.6257 3.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -7.0977 3.4382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -7.5243 3.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -7.4789 2.6207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -6.4228 3.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -7.9962 3.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -8.0425 3.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -8.5236 4.1127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -8.9584 3.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -8.9122 3.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -8.4310 3.0550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.6824 2.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -6.5084 1.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -9.3886 2.8806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.9638 1.0736 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 -3.7020 1.8865 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -4.3644 2.3290 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -4.7816 2.9115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -4.9096 2.1863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.2514 1.7212 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 -3.3604 0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.2088 1.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.0234 3.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.4389 1.1619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.0357 0.5860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.1628 -0.1349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.8064 -0.3692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.8446 -0.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -5.7612 1.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -5.3459 0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -7.5616 1.7672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -8.5439 1.5800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -9.6504 4.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -9.7223 4.8693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 0 0 0 2 3 2 0 0 0 0 3 4 1 0 0 0 0 4 5 2 0 0 0 0 5 6 1 0 0 0 0 6 1 2 0 0 0 0 4 7 1 0 0 0 0 7 8 1 0 0 0 0 8 9 2 0 0 0 0 9 10 1 0 0 0 0 10 5 1 0 0 0 0 7 11 2 0 0 0 0 9 12 1 0 0 0 0 12 13 2 0 0 0 0 13 14 1 0 0 0 0 14 15 2 0 0 0 0 15 16 1 0 0 0 0 16 17 2 0 0 0 0 17 12 1 0 0 0 0 3 18 1 0 0 0 0 1 19 1 0 0 0 0 20 16 1 0 0 0 0 21 22 1 1 0 0 0 22 23 1 1 0 0 0 24 23 1 1 0 0 0 24 25 1 0 0 0 0 25 26 1 0 0 0 0 26 21 1 0 0 0 0 21 27 1 0 0 0 0 22 28 1 0 0 0 0 23 29 1 0 0 0 0 26 30 1 0 0 0 0 24 18 1 0 0 0 0 30 31 1 0 0 0 0 31 32 1 0 0 0 0 32 33 1 0 0 0 0 32 34 2 0 0 0 0 2 35 1 0 0 0 0 35 36 1 0 0 0 0 6 37 1 0 0 0 0 37 38 1 0 0 0 0 15 39 1 0 0 0 0 39 40 1 0 0 0 0 M STY 1 3 SUP M SLB 1 3 3 M SAL 3 2 39 40 M SBL 3 1 42 M SMT 3 OCH3 M SVB 3 42 3.1301 1.8019 M STY 1 2 SUP M SLB 1 2 2 M SAL 2 2 37 38 M SBL 2 1 40 M SMT 2 OCH3 M SVB 2 40 0.0642 1.3987 M STY 1 1 SUP M SLB 1 1 1 M SAL 1 2 35 36 M SBL 1 1 38 M SMT 1 OCH3 M SVB 1 38 -1.3518 0.1923 S SKP 8 ID FL3FGCGS0004 KNApSAcK_ID C00004440 NAME 5,7,3'-Trihydroxy-6.8,4'-trimethoxyflavone 5-(6''-acetylglucoside) CAS_RN 72209-43-9 FORMULA C26H28O14 EXACTMASS 564.147905604 AVERAGEMASS 564.49212 SMILES O=C(c23)C=C(Oc2c(c(O)c(OC)c3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4COC(C)=O)O)OC)c(c1)ccc(c1O)OC M END