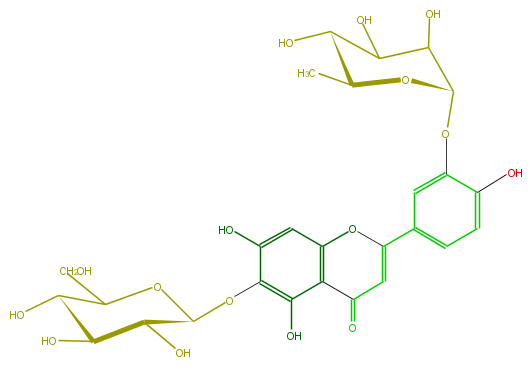
Mol:FL3FECGS0015
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.1155 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1155 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1155 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1155 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5010 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5010 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1174 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1174 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1174 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1174 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5010 -0.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5010 -0.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7338 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7338 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3502 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3502 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3502 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3502 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7338 -0.9150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7338 -0.9150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7338 -2.8935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7338 -2.8935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9664 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9664 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5946 -1.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5946 -1.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2228 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2228 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2228 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2228 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5946 0.1731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5946 0.1731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9664 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9664 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5946 0.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5946 0.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9072 0.2127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9072 0.2127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5010 -3.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5010 -3.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7306 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7306 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7306 -2.3377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7306 -2.3377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1616 -2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1616 -2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4570 -3.1819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4570 -3.1819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4423 -2.7873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4423 -2.7873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4631 -2.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4631 -2.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1747 -2.0652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1747 -2.0652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2111 -2.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2111 -2.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9072 -2.6242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9072 -2.6242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2653 -3.1542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2653 -3.1542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7344 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7344 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2738 3.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2738 3.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7137 1.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7137 1.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7959 2.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7959 2.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7444 1.8264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7444 1.8264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2413 2.6980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2413 2.6980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2627 2.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2627 2.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4641 2.8223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4641 2.8223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9289 2.2236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9289 2.2236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2702 3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2702 3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8837 3.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8837 3.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0640 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0640 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0877 -2.3027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0877 -2.3027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 2 22 1 0 0 0 0 | + | 2 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 22 1 0 0 0 0 | + | 26 22 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 35 18 1 0 0 0 0 | + | 35 18 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 28 42 1 0 0 0 0 | + | 28 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 47 0.8529 -0.6983 | + | M SBV 1 47 0.8529 -0.6983 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FECGS0015 | + | ID FL3FECGS0015 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES OC(C1O)C(O)C(Oc(c(O)5)c(O)c(c2c5)C(=O)C=C(c(c4)cc(c(O)c4)OC(C3O)OC(C(O)C3O)C)O2)OC1CO | + | SMILES OC(C1O)C(O)C(Oc(c(O)5)c(O)c(c2c5)C(=O)C=C(c(c4)cc(c(O)c4)OC(C3O)OC(C(O)C3O)C)O2)OC1CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.1155 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1155 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5010 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1174 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1174 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5010 -0.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7338 -2.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3502 -1.9827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3502 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7338 -0.9150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7338 -2.8935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9664 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5946 -1.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2228 -0.9151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2228 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5946 0.1731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9664 -0.1897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5946 0.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9072 0.2127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5010 -3.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7306 -0.9158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7306 -2.3377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1616 -2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4570 -3.1819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4423 -2.7873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4631 -2.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1747 -2.0652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2111 -2.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9072 -2.6242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2653 -3.1542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7344 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2738 3.0319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7137 1.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7959 2.0696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7444 1.8264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2413 2.6980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2627 2.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4641 2.8223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9289 2.2236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2702 3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8837 3.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0640 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0877 -2.3027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
19 15 1 0 0 0 0
3 20 1 0 0 0 0
1 21 1 0 0 0 0
2 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 22 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
37 41 1 0 0 0 0
35 18 1 0 0 0 0
42 43 1 0 0 0 0
28 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 ^CH2OH
M SBV 1 47 0.8529 -0.6983
S SKP 5
ID FL3FECGS0015
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES OC(C1O)C(O)C(Oc(c(O)5)c(O)c(c2c5)C(=O)C=C(c(c4)cc(c(O)c4)OC(C3O)OC(C(O)C3O)C)O2)OC1CO
M END