Mol:FL3FCFCS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1371 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1371 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1371 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1371 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5808 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5808 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0245 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0245 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0245 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0245 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5808 0.1560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5808 0.1560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5318 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5318 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0881 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0881 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0881 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0881 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5318 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5318 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5318 -1.6295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5318 -1.6295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5808 -1.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5808 -1.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7680 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7680 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3542 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3542 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9404 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9404 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3542 1.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3542 1.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7680 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7680 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6905 -1.0520 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.6905 -1.0520 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.1748 -1.6295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.1748 -1.6295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.6592 -1.3202 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.6592 -1.3202 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9580 -1.4027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9580 -1.4027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4530 -0.9489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4530 -0.9489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0098 -1.2170 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.0098 -1.2170 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.2407 -1.3697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2407 -1.3697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8172 -1.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8172 -1.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3498 -1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3498 -1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6471 1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6471 1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0969 2.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0969 2.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4943 0.4536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4943 0.4536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9942 1.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9942 1.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9404 0.9359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9404 0.9359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1917 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1917 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1501 -0.1682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1501 -0.1682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 2 22 1 0 0 0 0 | + | 2 22 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 17 28 1 0 0 0 0 | + | 17 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 16 32 1 0 0 0 0 | + | 16 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 24 34 1 0 0 0 0 | + | 24 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 34 35 | + | M SAL 4 2 34 35 |
| − | M SBL 4 1 37 | + | M SBL 4 1 37 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 37 -3.1917 -0.4538 | + | M SVB 4 37 -3.1917 -0.4538 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 32 33 | + | M SAL 3 2 32 33 |
| − | M SBL 3 1 35 | + | M SBL 3 1 35 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 35 3.5262 1.2742 | + | M SVB 3 35 3.5262 1.2742 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 30 31 | + | M SAL 2 2 30 31 |
| − | M SBL 2 1 33 | + | M SBL 2 1 33 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 33 -1.4943 0.4536 | + | M SVB 2 33 -1.4943 0.4536 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 28 29 | + | M SAL 1 2 28 29 |
| − | M SBL 1 1 31 | + | M SBL 1 1 31 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 31 2.6471 1.856 | + | M SVB 1 31 2.6471 1.856 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FCFCS0001 | + | ID FL3FCFCS0001 |
| − | KNApSAcK_ID C00006159 | + | KNApSAcK_ID C00006159 |
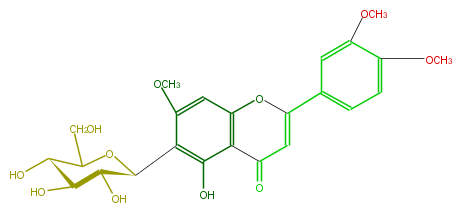
| − | NAME Isoorientin 7,3',4'-trimethyl ether;3',4',7-Tri-O-methylisoorientin;2-(3,4-Dimethoxyphenyl)-6-beta-D-glucopyranosyl-5-hydroxy-7-methoxy-4H-1-benzopyran-4-one | + | NAME Isoorientin 7,3',4'-trimethyl ether;3',4',7-Tri-O-methylisoorientin;2-(3,4-Dimethoxyphenyl)-6-beta-D-glucopyranosyl-5-hydroxy-7-methoxy-4H-1-benzopyran-4-one |
| − | CAS_RN 4261-30-7 | + | CAS_RN 4261-30-7 |
| − | FORMULA C24H26O11 | + | FORMULA C24H26O11 |
| − | EXACTMASS 490.147511674 | + | EXACTMASS 490.147511674 |
| − | AVERAGEMASS 490.45664 | + | AVERAGEMASS 490.45664 |
| − | SMILES O(C=1c(c4)cc(OC)c(c4)OC)c(c3)c(c(O)c(c3OC)[C@@H]([C@@H](O)2)OC([C@@H]([C@@H]2O)O)CO)C(C1)=O | + | SMILES O(C=1c(c4)cc(OC)c(c4)OC)c(c3)c(c(O)c(c3OC)[C@@H]([C@@H](O)2)OC([C@@H]([C@@H]2O)O)CO)C(C1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-1.1371 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1371 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5808 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0245 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0245 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5808 0.1560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5318 -1.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0881 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0881 -0.1652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5318 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5318 -1.6295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5808 -1.7708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7680 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3542 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9404 0.2590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3542 1.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7680 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6905 -1.0520 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.1748 -1.6295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6592 -1.3202 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9580 -1.4027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4530 -0.9489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0098 -1.2170 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.2407 -1.3697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8172 -1.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3498 -1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6471 1.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0969 2.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4943 0.4536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9942 1.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9404 0.9359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9404 0.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1917 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1501 -0.1682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
2 22 1 0 0 0 0
21 27 1 0 0 0 0
17 28 1 0 0 0 0
28 29 1 0 0 0 0
1 30 1 0 0 0 0
30 31 1 0 0 0 0
16 32 1 0 0 0 0
32 33 1 0 0 0 0
24 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 34 35
M SBL 4 1 37
M SMT 4 CH2OH
M SVB 4 37 -3.1917 -0.4538
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 32 33
M SBL 3 1 35
M SMT 3 OCH3
M SVB 3 35 3.5262 1.2742
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 30 31
M SBL 2 1 33
M SMT 2 OCH3
M SVB 2 33 -1.4943 0.4536
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 31
M SMT 1 OCH3
M SVB 1 31 2.6471 1.856
S SKP 8
ID FL3FCFCS0001
KNApSAcK_ID C00006159
NAME Isoorientin 7,3',4'-trimethyl ether;3',4',7-Tri-O-methylisoorientin;2-(3,4-Dimethoxyphenyl)-6-beta-D-glucopyranosyl-5-hydroxy-7-methoxy-4H-1-benzopyran-4-one
CAS_RN 4261-30-7
FORMULA C24H26O11
EXACTMASS 490.147511674
AVERAGEMASS 490.45664
SMILES O(C=1c(c4)cc(OC)c(c4)OC)c(c3)c(c(O)c(c3OC)[C@@H]([C@@H](O)2)OC([C@@H]([C@@H]2O)O)CO)C(C1)=O
M END