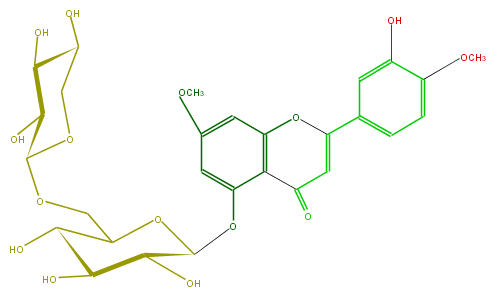
Mol:FL3FCEGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3819 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3819 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3819 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3819 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6828 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6828 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0166 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0166 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0166 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0166 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6828 0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6828 0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7159 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7159 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4151 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4151 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4151 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4151 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7159 0.6799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7159 0.6799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9821 -1.5054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9821 -1.5054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1141 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1141 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8269 0.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8269 0.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5396 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5396 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5396 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5396 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8269 1.9143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8269 1.9143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1141 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1141 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8269 2.8502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8269 2.8502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6828 -1.7409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6828 -1.7409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6172 -1.7896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6172 -1.7896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8178 -2.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8178 -2.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6667 -2.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6667 -2.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5561 -2.3852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5561 -2.3852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3632 -1.5780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3632 -1.5780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3703 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3703 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4336 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4336 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6913 -2.9068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6913 -2.9068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6552 -3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6552 -3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9172 -1.4671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9172 -1.4671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1331 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1331 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0981 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0981 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9191 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9191 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2832 -0.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2832 -0.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3180 0.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3180 0.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4972 1.2405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4972 1.2405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3277 3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3277 3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0328 2.5888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0328 2.5888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5623 0.2055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5623 0.2055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9736 -1.1791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9736 -1.1791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9359 1.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9359 1.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7112 2.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7112 2.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4546 2.0310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4546 2.0310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5623 1.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5623 1.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 29 39 1 0 0 0 0 | + | 29 39 1 0 0 0 0 |
| − | 39 33 1 0 0 0 0 | + | 39 33 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 42 1 0 0 0 0 | + | 15 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 45 0.5539 -0.9592 | + | M SBV 1 45 0.5539 -0.9592 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -0.9150 -0.5283 | + | M SBV 2 47 -0.9150 -0.5283 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FCEGS0002 | + | ID FL3FCEGS0002 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES COc(c3)cc(O1)c(c3OC(C(O)5)OC(C(O)C(O)5)COC(O4)C(O)C(O)C(C4)O)C(C=C(c(c2)ccc(c2O)OC)1)=O | + | SMILES COc(c3)cc(O1)c(c3OC(C(O)5)OC(C(O)C(O)5)COC(O4)C(O)C(O)C(C4)O)C(C=C(c(c2)ccc(c2O)OC)1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.3819 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3819 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6828 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0166 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0166 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6828 0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7159 -0.9350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4151 -0.5313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4151 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7159 0.6799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9821 -1.5054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1141 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8269 0.2683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5396 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5396 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8269 1.9143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1141 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8269 2.8502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6828 -1.7409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6172 -1.7896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8178 -2.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6667 -2.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5561 -2.3852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3632 -1.5780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3703 -2.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4336 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6913 -2.9068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6552 -3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9172 -1.4671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1331 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0981 1.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9191 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2832 -0.2594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3180 0.1906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4972 1.2405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3277 3.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0328 2.5888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5623 0.2055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9736 -1.1791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9359 1.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7112 2.5779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4546 2.0310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5623 1.3915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
3 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
25 29 1 0 0 0 0
23 19 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
29 39 1 0 0 0 0
39 33 1 0 0 0 0
40 41 1 0 0 0 0
1 40 1 0 0 0 0
42 43 1 0 0 0 0
15 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ OCH3
M SBV 1 45 0.5539 -0.9592
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -0.9150 -0.5283
S SKP 5
ID FL3FCEGS0002
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES COc(c3)cc(O1)c(c3OC(C(O)5)OC(C(O)C(O)5)COC(O4)C(O)C(O)C(C4)O)C(C=C(c(c2)ccc(c2O)OC)1)=O
M END