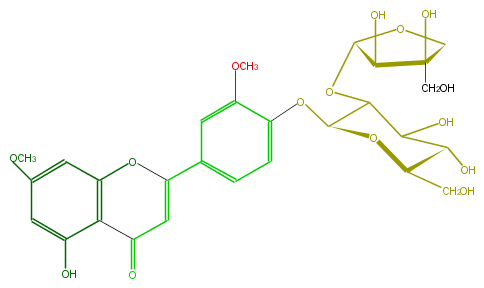
Mol:FL3FCDGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9152 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9152 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9152 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9152 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2142 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2142 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5134 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5134 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5134 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5134 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2142 -0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2142 -0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8124 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8124 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1114 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1114 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1114 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1114 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8124 -0.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8124 -0.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8124 -2.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8124 -2.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4107 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4107 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3036 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3036 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0180 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0180 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0180 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0180 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3036 0.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3036 0.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4107 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4107 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6630 0.7068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6630 0.7068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2142 -2.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2142 -2.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0680 0.6718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0680 0.6718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2333 0.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2333 0.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1683 -0.0441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1683 -0.0441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8433 -0.7397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8433 -0.7397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6781 -0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6781 -0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7431 -0.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7431 -0.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2533 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2533 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6037 0.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6037 0.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0574 -0.6973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0574 -0.6973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7561 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7561 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1892 1.4493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1892 1.4493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2411 1.5112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2411 1.5112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6536 1.8825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6536 1.8825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7255 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7255 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1892 2.5030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1892 2.5030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2411 2.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2411 2.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3036 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3036 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9072 2.8826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9072 2.8826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3959 -0.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3959 -0.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1728 0.8834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1728 0.8834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6600 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6600 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1728 -2.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1728 -2.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2411 0.9956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2411 0.9956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0812 1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0812 1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 15 1 0 0 0 0 | + | 18 15 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 18 21 1 0 0 0 0 | + | 18 21 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 29 1 0 0 0 0 | + | 33 29 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 29 26 1 0 0 0 0 | + | 29 26 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 16 36 1 0 0 0 0 | + | 16 36 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 1 38 1 0 0 0 0 | + | 1 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 31 42 1 0 0 0 0 | + | 31 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 36 37 | + | M SAL 1 2 36 37 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 41 0.0000 -0.7519 | + | M SBV 1 41 0.0000 -0.7519 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 38 39 | + | M SAL 2 2 38 39 |
| − | M SBL 2 1 43 | + | M SBL 2 1 43 |
| − | M SMT 2 ^ OCH3 | + | M SMT 2 ^ OCH3 |
| − | M SBV 2 43 0.4807 -0.4807 | + | M SBV 2 43 0.4807 -0.4807 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 40 41 | + | M SAL 3 2 40 41 |
| − | M SBL 3 1 45 | + | M SBL 3 1 45 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 45 -0.8167 0.3947 | + | M SBV 3 45 -0.8167 0.3947 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 42 43 | + | M SAL 4 2 42 43 |
| − | M SBL 4 1 47 | + | M SBL 4 1 47 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SBV 4 47 0.0000 0.5156 | + | M SBV 4 47 0.0000 0.5156 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FCDGS0004 | + | ID FL3FCDGS0004 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES O(c(c(OC)3)ccc(C(=C5)Oc(c4)c(C5=O)c(O)cc(OC)4)c3)C(C1OC(C(O)2)OCC2(CO)O)OC(CO)C(C(O)1)O | + | SMILES O(c(c(OC)3)ccc(C(=C5)Oc(c4)c(C5=O)c(O)cc(OC)4)c3)C(C1OC(C(O)2)OCC2(CO)O)OC(CO)C(C(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.9152 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9152 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2142 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5134 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5134 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2142 -0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8124 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1114 -1.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1114 -0.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8124 -0.5386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8124 -2.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4107 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3036 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0180 -0.5387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0180 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3036 0.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4107 0.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6630 0.7068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2142 -2.8631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0680 0.6718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2333 0.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1683 -0.0441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8433 -0.7397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6781 -0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7431 -0.0236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2533 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6037 0.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0574 -0.6973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7561 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1892 1.4493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2411 1.5112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6536 1.8825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7255 2.2125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1892 2.5030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2411 2.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3036 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9072 2.8826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3959 -0.4625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1728 0.8834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6600 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1728 -2.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2411 0.9956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0812 1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 15 1 0 0 0 0
3 19 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
18 21 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 29 1 0 0 0 0
30 34 1 0 0 0 0
31 35 1 0 0 0 0
29 26 1 0 0 0 0
36 37 1 0 0 0 0
16 36 1 0 0 0 0
38 39 1 0 0 0 0
1 38 1 0 0 0 0
40 41 1 0 0 0 0
23 40 1 0 0 0 0
42 43 1 0 0 0 0
31 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 36 37
M SBL 1 1 41
M SMT 1 OCH3
M SBV 1 41 0.0000 -0.7519
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 38 39
M SBL 2 1 43
M SMT 2 ^ OCH3
M SBV 2 43 0.4807 -0.4807
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 40 41
M SBL 3 1 45
M SMT 3 CH2OH
M SBV 3 45 -0.8167 0.3947
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 42 43
M SBL 4 1 47
M SMT 4 CH2OH
M SBV 4 47 0.0000 0.5156
S SKP 5
ID FL3FCDGS0004
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES O(c(c(OC)3)ccc(C(=C5)Oc(c4)c(C5=O)c(O)cc(OC)4)c3)C(C1OC(C(O)2)OCC2(CO)O)OC(CO)C(C(O)1)O
M END