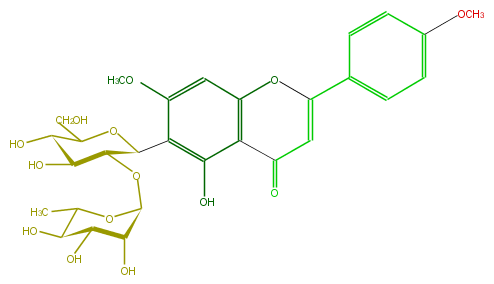
Mol:FL3FCBCS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.7790 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7790 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7790 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7790 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0642 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0642 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3495 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3495 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3495 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3495 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0642 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0642 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3653 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3653 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0800 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0800 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0800 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0800 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3653 1.2644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3653 1.2644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3653 -1.0297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3653 -1.0297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0642 -1.2112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0642 -1.2112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8739 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8739 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6273 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6273 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3804 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3804 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3804 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3804 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6273 2.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6273 2.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8739 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8739 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1479 0.1155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1479 0.1155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7029 -0.4719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7029 -0.4719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0623 -0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0623 -0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3947 -0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3947 -0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8934 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8934 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5477 -0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5477 -0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7584 -0.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7584 -0.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3777 -0.4668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3777 -0.4668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4257 -0.6753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4257 -0.6753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6234 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6234 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9578 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9578 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3146 -1.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3146 -1.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6676 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6676 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3331 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3331 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9763 -1.5472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9763 -1.5472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2639 -1.4194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2639 -1.4194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5018 -1.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5018 -1.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7586 -2.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7586 -2.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6670 -2.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6670 -2.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0904 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0904 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0511 0.8909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0511 0.8909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1331 2.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1331 2.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0511 2.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0511 2.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4429 1.2535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4429 1.2535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0855 2.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0855 2.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 9 1 0 0 0 0 | + | 13 9 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 22 2 1 0 0 0 0 | + | 22 2 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 24 38 1 0 0 0 0 | + | 24 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 1 42 1 0 0 0 0 | + | 1 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 43 0.5427 -0.4446 | + | M SBV 1 43 0.5427 -0.4446 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 45 -0.7527 -0.4345 | + | M SBV 2 45 -0.7527 -0.4345 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 ^ OCH3 | + | M SMT 3 ^ OCH3 |
| − | M SBV 3 47 0.6639 -0.4017 | + | M SBV 3 47 0.6639 -0.4017 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FCBCS0005 | + | ID FL3FCBCS0005 |
| − | FORMULA C29H34O14 | + | FORMULA C29H34O14 |
| − | EXACTMASS 606.194855796 | + | EXACTMASS 606.194855796 |
| − | AVERAGEMASS 606.57186 | + | AVERAGEMASS 606.57186 |
| − | SMILES O(C)c(c1)ccc(C(=C2)Oc(c5)c(c(c(c5OC)C(C3OC(O4)C(O)C(O)C(C(C)4)O)OC(C(O)C3O)CO)O)C2=O)c1 | + | SMILES O(C)c(c1)ccc(C(=C2)Oc(c5)c(c(c(c5OC)C(C3OC(O4)C(O)C(O)C(C(C)4)O)OC(C(O)C3O)CO)O)C2=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.7790 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7790 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0642 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3495 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3495 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0642 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3653 -0.3862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0800 0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0800 0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3653 1.2644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3653 -1.0297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0642 -1.2112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8739 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6273 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3804 1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3804 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6273 2.5954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8739 2.1606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1479 0.1155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7029 -0.4719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0623 -0.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3947 -0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8934 0.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5477 -0.0017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7584 -0.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3777 -0.4668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4257 -0.6753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6234 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9578 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3146 -1.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6676 -1.9427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3331 -1.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9763 -1.5472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2639 -1.4194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5018 -1.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7586 -2.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6670 -2.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0904 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0511 0.8909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1331 2.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0511 2.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4429 1.2535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0855 2.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
13 9 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 27 1 0 0 0 0
22 2 1 0 0 0 0
38 39 1 0 0 0 0
24 38 1 0 0 0 0
40 41 1 0 0 0 0
16 40 1 0 0 0 0
42 43 1 0 0 0 0
1 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 ^ CH2OH
M SBV 1 43 0.5427 -0.4446
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 OCH3
M SBV 2 45 -0.7527 -0.4345
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 47
M SMT 3 ^ OCH3
M SBV 3 47 0.6639 -0.4017
S SKP 5
ID FL3FCBCS0005
FORMULA C29H34O14
EXACTMASS 606.194855796
AVERAGEMASS 606.57186
SMILES O(C)c(c1)ccc(C(=C2)Oc(c5)c(c(c(c5OC)C(C3OC(O4)C(O)C(O)C(C(C)4)O)OC(C(O)C3O)CO)O)C2=O)c1
M END