Mol:FL3FCADS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3766 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3766 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3766 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3766 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2640 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2640 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2640 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2640 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 -0.2057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 -0.2057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2923 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2923 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8486 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8486 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8486 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8486 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2923 -0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2923 -0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2923 -1.9913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2923 -1.9913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5956 2.2725 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.5956 2.2725 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.2012 1.8137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2012 1.8137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9442 1.1531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.9442 1.1531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9374 0.5156 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.9374 0.5156 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.4740 0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4740 0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7791 1.5568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.7791 1.5568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.8504 3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8504 3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2360 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2360 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5797 0.7745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5797 0.7745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 -2.1326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 -2.1326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5284 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5284 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1146 -0.4412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1146 -0.4412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7008 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7008 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7008 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7008 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1146 0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1146 0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5284 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5284 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2867 0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2867 0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7729 -2.4274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.7729 -2.4274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.4017 -2.9174 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4017 -2.9174 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8672 -2.7095 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8672 -2.7095 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.3514 -2.7039 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.3514 -2.7039 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7262 -2.3291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7262 -2.3291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1938 -2.5759 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.1938 -2.5759 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.3638 -2.3757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3638 -2.3757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7521 -3.3786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7521 -3.3786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5609 -3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5609 -3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0832 1.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0832 1.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6313 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6313 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7339 0.0918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7339 0.0918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2340 0.9578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2340 0.9578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8864 -2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8864 -2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4698 -1.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4698 -1.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 12 13 1 1 0 0 0 | + | 12 13 1 1 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 15 14 1 1 0 0 0 | + | 15 14 1 1 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 12 18 1 0 0 0 0 | + | 12 18 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 6 15 1 0 0 0 0 | + | 6 15 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 9 22 1 0 0 0 0 | + | 9 22 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 21 1 0 0 0 0 | + | 32 21 1 0 0 0 0 |
| − | 17 38 1 0 0 0 0 | + | 17 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 46 -2.5923 -1.7917 | + | M SVB 3 46 -2.5923 -1.7917 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 38 39 | + | M SAL 2 2 38 39 |
| − | M SBL 2 1 42 | + | M SBL 2 1 42 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 42 -0.0832 1.9585 | + | M SVB 2 42 -0.0832 1.9585 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 44 -1.7339 0.0918 | + | M SVB 1 44 -1.7339 0.0918 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FCADS0004 | + | ID FL3FCADS0004 |
| − | KNApSAcK_ID C00006277 | + | KNApSAcK_ID C00006277 |
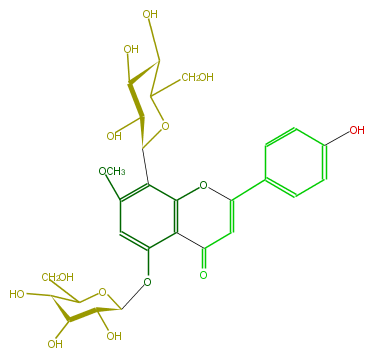
| − | NAME Isoswertisin 5-O-glucoside | + | NAME Isoswertisin 5-O-glucoside |
| − | CAS_RN 73051-76-0 | + | CAS_RN 73051-76-0 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES O[C@H]([C@H](O)1)C(O[C@@H](Oc(c34)cc(c(c3OC(c(c5)ccc(c5)O)=CC(=O)4)[C@H](O2)[C@@H](O)[C@@H](O)[C@H](C2CO)O)OC)[C@H]1O)CO | + | SMILES O[C@H]([C@H](O)1)C(O[C@@H](Oc(c34)cc(c(c3OC(c(c5)ccc(c5)O)=CC(=O)4)[C@H](O2)[C@@H](O)[C@@H](O)[C@H](C2CO)O)OC)[C@H]1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.3766 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3766 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2640 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2640 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 -0.2057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2923 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8486 -1.1693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8486 -0.5269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2923 -0.2057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2923 -1.9913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5956 2.2725 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.2012 1.8137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9442 1.1531 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9374 0.5156 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4740 0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7791 1.5568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.8504 3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2360 2.5289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5797 0.7745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 -2.1326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5284 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1146 -0.4412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7008 -0.1027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7008 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1146 0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5284 0.5742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2867 0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7729 -2.4274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.4017 -2.9174 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8672 -2.7095 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3514 -2.7039 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7262 -2.3291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1938 -2.5759 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.3638 -2.3757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7521 -3.3786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5609 -3.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0832 1.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6313 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7339 0.0918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2340 0.9578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8864 -2.0336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4698 -1.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
12 13 1 1 0 0 0
13 14 1 1 0 0 0
15 14 1 1 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 12 1 0 0 0 0
12 18 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
6 15 1 0 0 0 0
3 21 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
9 22 1 0 0 0 0
25 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 21 1 0 0 0 0
17 38 1 0 0 0 0
38 39 1 0 0 0 0
1 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 46
M SMT 3 CH2OH
M SVB 3 46 -2.5923 -1.7917
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 38 39
M SBL 2 1 42
M SMT 2 CH2OH
M SVB 2 42 -0.0832 1.9585
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SVB 1 44 -1.7339 0.0918
S SKP 8
ID FL3FCADS0004
KNApSAcK_ID C00006277
NAME Isoswertisin 5-O-glucoside
CAS_RN 73051-76-0
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES O[C@H]([C@H](O)1)C(O[C@@H](Oc(c34)cc(c(c3OC(c(c5)ccc(c5)O)=CC(=O)4)[C@H](O2)[C@@H](O)[C@@H](O)[C@H](C2CO)O)OC)[C@H]1O)CO
M END