Mol:FL3FAEGS0011
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6474 0.4808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6474 0.4808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6474 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6474 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0514 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0514 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7500 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7500 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7500 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7500 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0514 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0514 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4488 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4488 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1477 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1477 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1477 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1477 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4488 0.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4488 0.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4488 -1.4224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4488 -1.4224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8462 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8462 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5583 0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5583 0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2704 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2704 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2704 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2704 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5583 2.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5583 2.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8462 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8462 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0514 -1.5984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0514 -1.5984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3619 0.8934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3619 0.8934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2800 1.1821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2800 1.1821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4813 0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4813 0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3311 0.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3311 0.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2212 0.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2212 0.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0278 1.3937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0278 1.3937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2026 0.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2026 0.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0892 0.8302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0892 0.8302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5189 0.2513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5189 0.2513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3614 -0.0584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3614 -0.0584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5644 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5644 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1649 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1649 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0101 -1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0101 -1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8482 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8482 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2476 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2476 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4025 -1.2541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4025 -1.2541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5837 -1.0821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5837 -1.0821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7887 -1.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7887 -1.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4368 -2.4471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4368 -2.4471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7644 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7644 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5583 2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5583 2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0098 1.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0098 1.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9030 1.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9030 1.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9824 2.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9824 2.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0892 1.4146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0892 1.4146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 42 1 0 0 0 0 | + | 15 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.8072 -0.6927 | + | M SBV 1 45 0.8072 -0.6927 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -0.7120 -0.4110 | + | M SBV 2 47 -0.7120 -0.4110 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAEGS0011 | + | ID FL3FAEGS0011 |
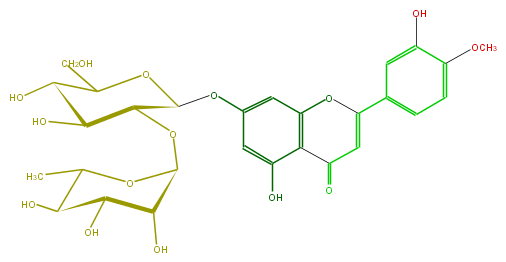
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES C(C(O)4)(C(OC(O5)C(O)C(C(C5C)O)O)C(OC4CO)Oc(c1)cc(O2)c(C(=O)C=C2c(c3)ccc(c3O)OC)c1O)O | + | SMILES C(C(O)4)(C(OC(O5)C(O)C(C(C5C)O)O)C(OC4CO)Oc(c1)cc(O2)c(C(=O)C=C2c(c3)ccc(c3O)OC)c1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.6474 0.4808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6474 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0514 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7500 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7500 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0514 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4488 -0.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1477 -0.3899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1477 0.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4488 0.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4488 -1.4224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8462 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5583 0.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2704 0.8202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2704 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5583 2.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8462 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0514 -1.5984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3619 0.8934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2800 1.1821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4813 0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3311 0.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2212 0.5871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0278 1.3937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2026 0.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0892 0.8302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5189 0.2513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3614 -0.0584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5644 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1649 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0101 -1.6343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8482 -1.9644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2476 -0.9243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4025 -1.2541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5837 -1.0821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7887 -1.7650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4368 -2.4471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7644 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5583 2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0098 1.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9030 1.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9824 2.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0892 1.4146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 28 1 0 0 0 0
23 19 1 0 0 0 0
16 39 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 1 0 0 0 0
15 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.8072 -0.6927
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -0.7120 -0.4110
S SKP 5
ID FL3FAEGS0011
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES C(C(O)4)(C(OC(O5)C(O)C(C(C5C)O)O)C(OC4CO)Oc(c1)cc(O2)c(C(=O)C=C2c(c3)ccc(c3O)OC)c1O)O
M END