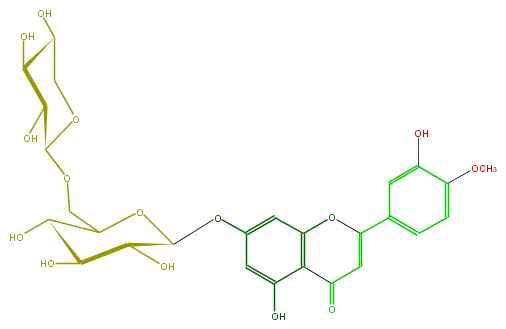
Mol:FL3FAEGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5818 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5818 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5818 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5818 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1191 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1191 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8201 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8201 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8201 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8201 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1191 -1.2926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1191 -1.2926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2219 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2219 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2219 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2219 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 -3.5424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 -3.5424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9226 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9226 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6369 -1.7051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6369 -1.7051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3515 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3515 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3515 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3515 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6369 -0.0553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6369 -0.0553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9226 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9226 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6369 0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6369 0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2814 -1.2933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2814 -1.2933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1191 -3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1191 -3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4469 -1.3609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4469 -1.3609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6457 -2.4186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6457 -2.4186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4919 -1.9699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4919 -1.9699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3786 -1.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3786 -1.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1876 -1.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1876 -1.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1971 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1971 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1370 -1.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1370 -1.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3796 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3796 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6022 -2.4836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6022 -2.4836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9283 -1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9283 -1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9851 -0.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9851 -0.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2849 3.1230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2849 3.1230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0397 2.3682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0397 2.3682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5110 1.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5110 1.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5110 0.3737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5110 0.3737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7562 1.1285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7562 1.1285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2849 2.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2849 2.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6289 3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6289 3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0397 3.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0397 3.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9632 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9632 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0269 -0.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0269 -0.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1370 -0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1370 -0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 31 30 1 0 0 0 0 | + | 31 30 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 15 41 1 0 0 0 0 | + | 15 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 46 -0.6754 -0.3899 | + | M SBV 1 46 -0.6754 -0.3899 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAEGS0005 | + | ID FL3FAEGS0005 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(O)(c13)cc(OC(C4O)OC(COC(O5)C(O)C(C(O)C5)O)C(O)C4O)cc(OC(=CC(=O)3)c(c2)cc(O)c(OC)c2)1 | + | SMILES c(O)(c13)cc(OC(C4O)OC(COC(O5)C(O)C(C(O)C5)O)C(O)C4O)cc(OC(=CC(=O)3)c(c2)cc(O)c(OC)c2)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.5818 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5818 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1191 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8201 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8201 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1191 -1.2926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 -2.9113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2219 -2.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2219 -1.6972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 -3.5424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9226 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6369 -1.7051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3515 -1.2927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3515 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6369 -0.0553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9226 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6369 0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2814 -1.2933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1191 -3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4469 -1.3609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6457 -2.4186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4919 -1.9699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3786 -1.9580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1876 -1.1487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1971 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1370 -1.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3796 -2.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6022 -2.4836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9283 -1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9851 -0.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2849 3.1230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0397 2.3682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5110 1.4410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5110 0.3737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7562 1.1285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2849 2.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6289 3.7189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0397 3.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9632 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0269 -0.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1370 -0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 19 1 0 0 0 0
26 30 1 0 0 0 0
31 30 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 31 1 0 0 0 0
41 42 1 0 0 0 0
15 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 OCH3
M SBV 1 46 -0.6754 -0.3899
S SKP 5
ID FL3FAEGS0005
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(O)(c13)cc(OC(C4O)OC(COC(O5)C(O)C(C(O)C5)O)C(O)C4O)cc(OC(=CC(=O)3)c(c2)cc(O)c(OC)c2)1
M END