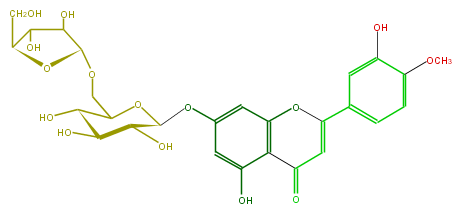
Mol:FL3FAEGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.2063 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2063 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2063 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2063 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4946 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4946 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1957 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1957 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1957 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1957 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4946 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4946 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8966 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8966 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5976 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5976 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5976 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5976 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8966 -0.1567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8966 -0.1567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8966 -2.5632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8966 -2.5632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2983 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2983 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0128 -0.5695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0128 -0.5695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7273 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7273 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7273 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7273 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0128 1.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0128 1.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2983 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2983 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0128 1.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0128 1.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9059 -0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9059 -0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4946 -2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4946 -2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7834 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7834 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2225 -0.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2225 -0.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4148 -0.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4148 -0.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6354 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6354 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2018 -0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2018 -0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9084 -0.4669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9084 -0.4669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5045 -0.4358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5045 -0.4358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0287 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0287 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5794 -1.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5794 -1.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4113 0.1001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4113 0.1001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1468 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1468 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0229 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0229 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4705 1.3704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4705 1.3704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5955 0.8543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5955 0.8543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6755 1.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6755 1.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0229 1.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0229 1.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1449 2.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1449 2.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4113 0.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4113 0.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4705 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4705 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5114 2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5114 2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4012 1.0571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4012 1.0571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5114 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5114 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 38 30 1 0 0 0 0 | + | 38 30 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 15 41 1 0 0 0 0 | + | 15 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 0.0000 -0.8972 | + | M SBV 1 44 0.0000 -0.8972 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 46 -0.6739 -0.3891 | + | M SBV 2 46 -0.6739 -0.3891 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAEGS0003 | + | ID FL3FAEGS0003 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES O(CC(O2)C(O)C(C(O)C(Oc(c5)cc(O)c(c45)C(=O)C=C(O4)c(c3)cc(O)c(OC)c3)2)O)C(C(O)1)OC(CO)C1O | + | SMILES O(CC(O2)C(O)C(C(O)C(Oc(c5)cc(O)c(c45)C(=O)C=C(O4)c(c3)cc(O)c(OC)c3)2)O)C(C(O)1)OC(CO)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.2063 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2063 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4946 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1957 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1957 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4946 -0.1567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8966 -1.7756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5976 -1.3709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5976 -0.5616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8966 -0.1567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8966 -2.5632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2983 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0128 -0.5695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7273 -0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7273 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0128 1.0805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2983 0.6680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0128 1.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9059 -0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4946 -2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7834 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2225 -0.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4148 -0.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6354 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2018 -0.0940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9084 -0.4669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5045 -0.4358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0287 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5794 -1.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4113 0.1001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1468 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0229 1.8415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4705 1.3704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5955 0.8543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6755 1.3255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0229 1.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1449 2.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4113 0.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4705 2.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.5114 2.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4012 1.0571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5114 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 19 1 0 0 0 0
26 30 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 31 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
35 38 1 0 0 0 0
38 30 1 0 0 0 0
39 40 1 0 0 0 0
33 39 1 0 0 0 0
41 42 1 0 0 0 0
15 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 0.0000 -0.8972
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 OCH3
M SBV 2 46 -0.6739 -0.3891
S SKP 5
ID FL3FAEGS0003
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES O(CC(O2)C(O)C(C(O)C(Oc(c5)cc(O)c(c45)C(=O)C=C(O4)c(c3)cc(O)c(OC)c3)2)O)C(C(O)1)OC(CO)C1O
M END